BACKGROUND
Infectious intestinal disease (IID) is common in childhood with each child probably experiencing four or more episodes before age 5 years, although not all lead to consultation with a general practitioner (GP). Rotaviruses, adenoviruses, astroviruses, noroviruses and sapoviruses are the major aetiological agents: bacterial and parasitic infections are much less frequent [Reference de Wit1, Reference de Wit2].
Electron microscopy has traditionally been used for the detection of enteric viruses but this, and other early methods are relatively insensitive. More recent molecular methods for the detection of enteric bacteria, parasites and viruses provide improved sensitivity, especially for enteric viruses [Reference Amar, Dear and McLauchlin3–Reference Amar5].
Although the association of these pathogens with IID is well understood, the burden of disease associated with the different organisms is poorly described. Current data on the burden of disease are mainly derived from routine voluntary reporting by clinical microbiology laboratories although a few structured surveillance studies have been undertaken [Reference Simpson6, Reference Iturriza7].
The aims of the present study were: (1) to establish a structured surveillance scheme to monitor the incidence of IID in pre-school children presenting to GPs; (2) to determine the burden of disease associated with rotavirus in pre-school children in order to inform policy on childhood rotavirus vaccination.
METHODS
Study design and denominator data collection
Sixty-five volunteer general practices were recruited, most of which belonged to the Royal College of General Practitioners Weekly Returns Service (RCGP WRS) [Reference Djuretic8]. The WRS is a sentinel GP surveillance scheme comprising 100 practices (500 GPs) in England and Wales and an average monitored population of 950 000 [9]. Additional practices (n=22) were recruited in some areas to achieve a larger number of potential samples and a broader national distribution. The study was conducted between February 2006 and September 2007, with each practice enrolling subjects for 12 months. The combined practice-registered population in age group 0–4 years provided the denominator for reporting the incidence of new episodes of IID on a weekly basis.
When a child aged <5 years presented with diarrhoea, diarrhoea and vomiting or other symptoms indicative of IID, the parent was asked to provide the next soiled nappy for the investigation of pathogens. The nappy and an accompanying specimen form were transported (daily on request) to the Health Protection Agency (HPA) laboratory in Colindale, London (Fig. 1). In recognition of their cooperation, a small teddy bear was given to each child after they had provided a nappy; a donation to the Save the Children Fund was made for each nappy collected.

Fig. 1. Flow diagram illustrating the recruitment of children, collection and processing of samples, and the communication of results during the Nappy Study.
Specimens were investigated within 24 h of receipt at the HPA in order to provide a rapid diagnostic service to the practices and patients involved. Results were reported to the parent of the child if this was indicated on the specimen form; all results were communicated to the practices by post within 24–48 working hours of specimen receipt. The detection of pathogens including Salmonella spp. and Cryptosporidium was communicated to the GP by telephone to facilitate improved patient management.
The practices (43/65) provided clinical incidence data as part of their routine twice-weekly contribution to the WRS. Data from the WRS covering 2001–2007 were used to estimate the burden of illness from the perspective of health service provision.
Ethical approval for the study was obtained from West Midlands Multi-Centre Research Ethics Committee (05/MRE07/52).
Laboratory investigation
Nucleic acid extraction and reverse transcription
Faecal samples were prepared as 10% suspensions in balanced salt solution (M199; Sigma, Dorset, UK) and stored at 4–8°C. RNA extraction and reverse transcription were performed as previously described [Reference Boom10, Reference Iturriza-Gómara11]. DNA was extracted from faecal suspension using a modified method of Boom et al. [Reference Boom10], incorporating mechanical disruption of cells using the MagNA Lyser instrument and ceramic beads (MagNA Lyser Green Beads; Roche Diagnostics, Mannheim, Germany) in order to extract DNA from parasites.
Real-time PCR was performed with the PRISM 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA); block-based PCR was also used, with visualization of PCR products through gel electrophoresis as previously described [Reference Amar4, Reference Boom10, Reference Iturriza-Gomara, Kang and Gray12]. RNAse-free sterile distilled water (Invitrogen, Paisley, UK) was used as a negative control throughout the study. Appropriate positive controls for each pathogen were also included in each batch of nucleic acid extractions and PCR procedures.
Viruses
Extracted DNA was used in real-time PCR detection of enteric adenovirus using oligonucleotide primers described previously [Reference Simpson6] in combination with a 3′-minor groove binder probe (5′FAM-TGCACCTCTTGGACTAGT-MGB3′; Applied Biosystems). Primers were used at 600 nm and probe at 100 nm concentrations using Platinum qPCR Mastermix (Invitrogen). The cycle conditions were as follows: 50°C for 2 min, then 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.
cDNA was used in the real-time PCR detection of norovirus [Reference Amar5, Reference Kageyama13] and rotavirus. A real-time group A rotavirus-specific PCR was designed using VP6-specific primers as previously described [Reference Amar4]. The assay was performed using Platinum qPCR Mastermix, 800 nm of each primer VP6-F and VP6-R and 100 nm of the Rota VP6 dual-labelled probe: 5′FAM-CCA CCR AAY ATG ACR CCA GCN GTA RMW GCA TTA TTT CC-Tamra3′. The cycle conditions were as follows: 50°C for 2 min, then 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Extracted cDNA was also used for the block-based PCR detection of astrovirus and sapovirus using oligonucleotide primers and methods described previously [Reference Simpson6].
Rotavirus-positive and norovirus-positive samples were genotyped using methods described previously [Reference Iturriza-Gomara, Kang and Gray12, Reference Gallimore14].
Bacteria and parasites
The real-time PCR for the detection of the bacterial pathogens, enteroaggregative Escherichia coli, Salmonella spp. and Campylobacter spp. and protozoa, Cryptosporidium spp. and Giardia spp. were performed using oligonucleotide primers and methods described previously [Reference Amar4, Reference Amar5].
RESULTS
Epidemiological data
The incidence of IID in the WRS between 2001 and 2007 is shown in Figure 2. The incidence and temporal distribution of IID in the 43 WRS practices recruited to the present study was similar to that reported by the total 100 WRS network (Fig. 3). Incidence of IID peaked in late winter (March/April). The 43 practices reported a total of 1584 new episodes of IID and from these 443 specimens were submitted for analysis; a specimen capture rate of 28%. The 22 additional practices provided 140 specimens. From the clinical data available (WRS practices) 443 nappies (267 positive for pathogens) were collected from an average weekly population (aged 0–4 years) of 19 774.
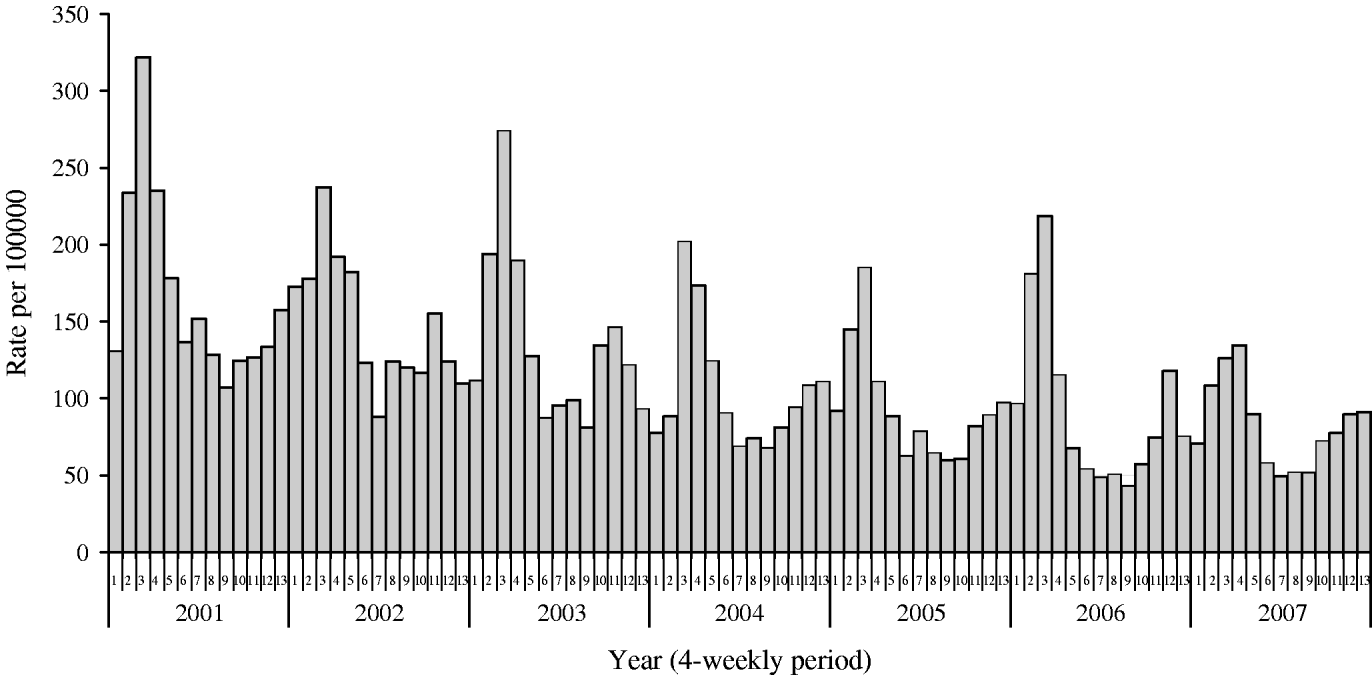
Fig. 2. Mean weekly incidence of infectious intestinal disease in 4-weekly periods for children aged 0–4 years.

Fig. 3. Clinical incidence of infectious intestinal disease in Weekly Returns Service (WRS) Nappy Study practices (![]() ) compared to WRS data (–––) for England and Wales over the same period for children aged 0–4 years.
) compared to WRS data (–––) for England and Wales over the same period for children aged 0–4 years.
A total of 583 soiled nappies were collected from 554 children; a significant pathogen was detected in 361 specimens (62% of the total). Norovirus (24·5%), rotavirus (19%) and sapovirus (12·7%) were the most frequently detected pathogens. Mixed infections were observed in 68 specimens (11·7%): two pathogens were found in 64 specimens and three in a further four specimens. Multiple infections consisted mostly of a combination of norovirus with one or more other pathogens (8·4%), followed by rotavirus (4·7%) and sapovirus (4·3%). Bacterial pathogens were detected as follows: enteroaggregative E. coli (1·2%); Campylobacter jejuni (1·0%); Campylobacter coli (0·3%); and Salmonella spp. (0·2%). Giardia was detected in one patient (0·2%); Cryptosporidium was not detected throughout the study. No pathogen was detected in 222 specimens tested (38%) but in June, July and August no pathogen was detected in >50% of specimens.
The majority of children presented with diarrhoea and vomiting (62·2%). Children with rotavirus, norovirus and adenovirus mostly presented with both symptoms (68·7%): children with astrovirus and sapovirus mainly presented solely with diarrhoea (52·8%).
More than one specimen was collected from 21 children (range 2–6 specimens). Subsequent specimens commonly produced a different virus or different genotype of the same virus (Table 1). The period between the first and second episode averaged 3·5 months (range 1–8 months) and did not differ significantly whether with the same or a different pathogen.
Table 1. Characterisation of multiple infections by patient, age of infection, pathogen and genotypic characterization
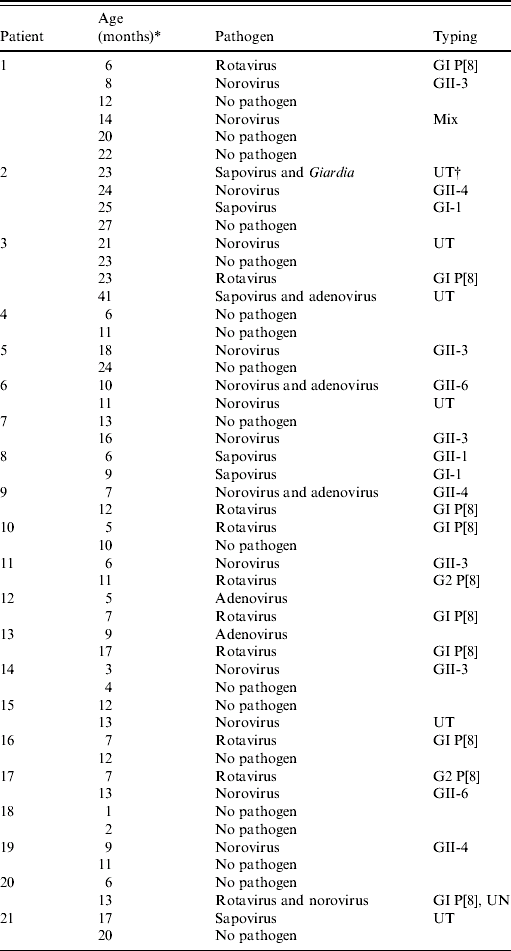
* Age at time of episode of infectious intestinal disease.
† UT, Untypable isolates (due to unsatisfactory quality of sequence trace data).
Rotavirus analysis
Rotavirus infections reported in our study were compared with those reported in the routine laboratory-based surveillance scheme of the HPA: both showed seasonal peaks in March 2006 and 2007 (Fig. 4). In the present study, no cases were detected between September and December in 2006 and none after July 2007 (Fig. 5). The highest percentage of rotavirus cases was in age group 19–24 months (26·7%) and lowest in 0–6 months (12·1%; Table 2).

Fig. 4. Temporal distribution of rotavirus infections between January and October 2006 and 2007 in England according passive surveillance (laboratory reports), and in the Nappy Study. Y axes scales indicate number.
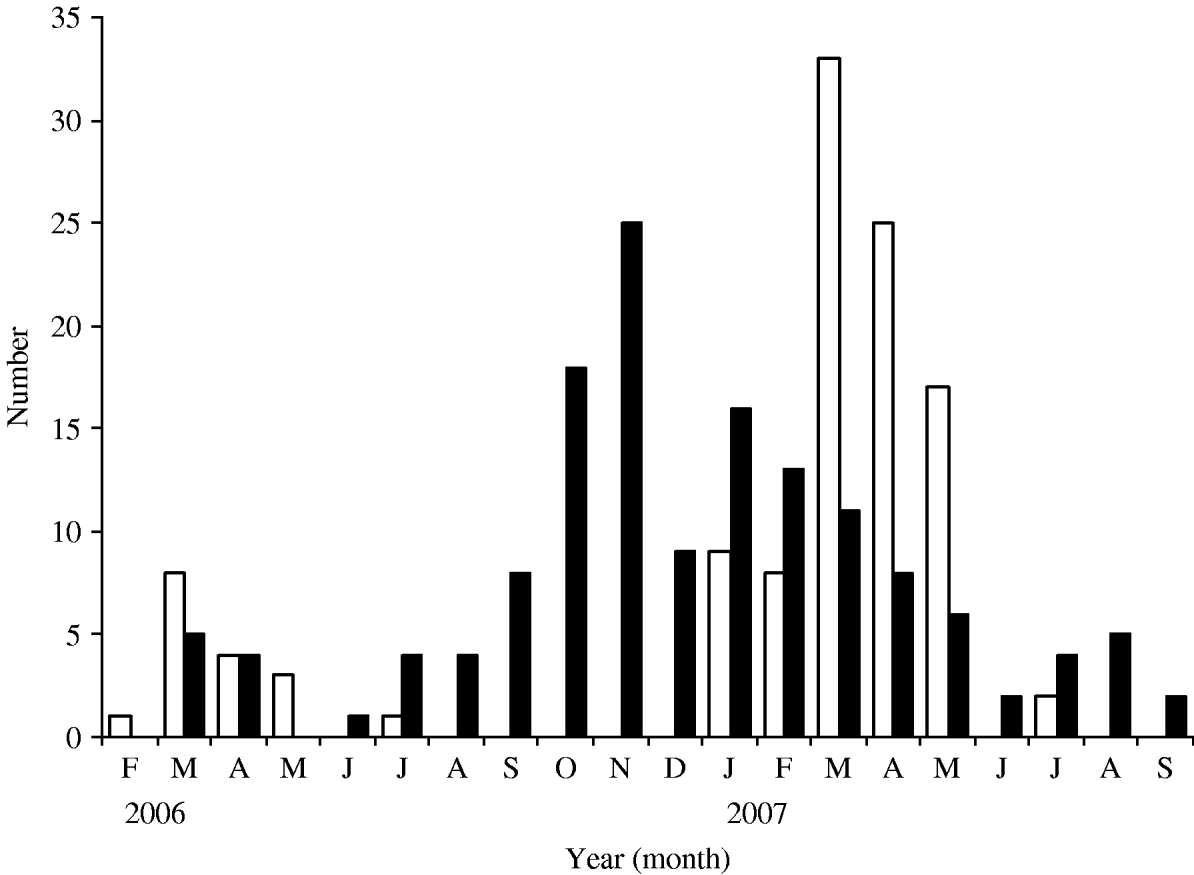
Fig. 5. Seasonal distribution of rotavirus (□) and norovirus (■) infections.
Table 2. Age-specific virology
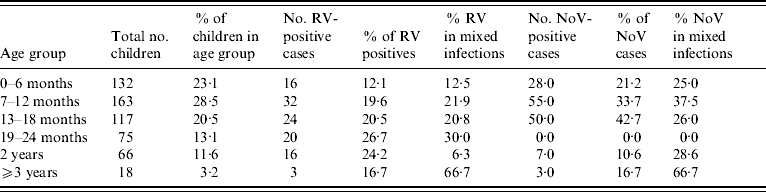
RV, Rotavirus; NoV, norovirus.
Rotavirus strain G1P[Reference Djuretic8] was most commonly detected (65·8%) followed by G2P[Reference Amar4] (8·1%) and G9P[Reference Djuretic8] (8·1%). Possible reassortant strains, G2P[Reference Djuretic8] and G4P[Reference Amar4], and a recently emerged strain G12P[Reference Djuretic8] represented 4·5%, 1·8% and 0·9% of the total strains, respectively (Fig. 6). The genotype distribution was similar to that seen in the HPA national rotavirus surveillance programme which included strains from 508 patients in the community (Fig. 6). These were similar in age and gender distribution and genotype and seasonality to those isolated from children admitted to hospital (data not shown).

Fig. 6. Rotavirus genotype distribution in the United Kingdom. □, Total; ■, community;![]() , Nappy Study.
, Nappy Study.
Norovirus analysis
Norovirus infections were seasonal and peaked in November. There were only two months (February and May 2006) when norovirus was not detected (Fig. 5). The incidence of norovirus gastroenteritis was significantly greater (P=0·02) in the 7–12 months age group (33·7%, Table 2) than in any other age group.
GII-3 was the most commonly detected norovirus strain (42·9%) followed by GII-4 (39·7%). Lower incidences of 1·7%, 5·8% and 1·7% were associated with GII-2, GII-6 and GII-7, respectively. GII-4 was detected in a further 1·7% of patients and mixed infections with more than one norovirus genotype in 6·6%. When compared with national data, the norovirus genotype distribution was significantly different (P<0·005 for both GII-3 and GII-4) to that of strains associated with outbreaks, predominantly occurring in hospitals and nursing homes. Only 6·3% of outbreaks in these settings were associated with GII-3 whereas 82·7% were caused by GII-4.
DISCUSSION
The present study has tested the feasibility of collecting soiled nappies directly for pathological investigation and is the first published report of this method. We achieved a good specimen capture rate (28%) similar to that found in most similar studies collecting faecal specimens by the traditional ‘spoon and pot’ [Reference de Wit1]. By linking the specimen capture to routine data collection on the incidence of IID we have shown, both in the clinical diagnosis and in the comparison with national laboratory surveillance, that our findings are consistent with the broader national picture.
The present study was designed to provide a diagnostic service to GPs and patients. It was therefore essential to examine the specimens for a broad range of pathogens and not simply focus on viruses of special interest. Furthermore, prompt analysis and reporting of results was fundamental. A primary purpose of our study was to establish a methodology which might allow us to monitor the impact of a rotavirus vaccine should vaccination become national policy. We believe our study has paved the way for a routine system of structured surveillance of IID in the community to establish the burden of disease in pre- and post-vaccination eras.
A total of 583 soiled nappies from children with clinically diagnosed IID were investigated. A pathogen was detected in 62% of the IID episodes; norovirus was identified in 25% of episodes, followed by rotavirus in 19% and sapovirus in 12%; bacterial and parasitic infections were rare.
Consecutive IID episodes were reported in 21/554 (3·8%) children and were associated with either re-infections with the same virus of a different genotype, with another pathogen, multiple pathogens or no aetiological agent was identified from the episode(s). The presence of multiple and consecutive infections with enteric pathogens is associated with the protection offered by mucosal immunity. Immunity tends to be homotypic and short-lived allowing re-infections and infections with closely related organisms.
The present study has shown that, at least during the period studied, norovirus was more common as a cause of IID in children than has been recognized hitherto [Reference de Wit2, Reference Iturriza7, Reference Svraka15]. Norovirus is the most common cause of outbreaks of diarrhoea in semi-closed communities (hospitals, cruise ships, etc.) [Reference Svraka15, Reference Gallimore, Richards and Gray16], but we demonstrated that in 2 two years of our study it was the commonest virus in community-dwelling children.
It is of interest to note that in the present study, reports of norovirus infection in children preceded the reports to the HPA of outbreaks of norovirus gastroenteritis, which were mainly from adults (Fig. 7). This may provide an early warning indicator of norovirus activity in older age groups giving hospitals and residential homes more time to prepare for, and better manage outbreaks in their facilities.

Fig. 7. Temporal distribution of norovirus infections in children and reported in the present study (–––) and of outbreaks reported in England and Wales for the same period (– – –). Y axes scales indicate number.
The seasonality of rotavirus and norovirus infections was predominantly in winter and early spring but the relatively high rates of nil detection of any pathogen in the high summer months are of interest. This would suggest that an aetiological agent associated with gastroenteritis in young children in the summer months has yet to be identified.
The significant difference in the distribution of norovirus genotypes within our study compared with reports of outbreaks in closed communities is similar to that reported previously. Gallimore et al. [Reference Gallimore17] reported significantly higher incidences of infection with GI and non-GII-4 strains in the community compared to nursing homes or hospitals. Although in the present study GI strains were uncommon, non-GII-4 strains were detected more frequently than GII-4 strains.
Rotavirus was, in the present study, the second most common cause of IID. The rotavirus season started and peaked later in 2007 than in previous seasons, but was not more prolonged into the spring months than in previous seasons. The data collected for England and Wales (HPA) and other European countries during 2007 suggested a decrease in the total number of reports of rotavirus infections (J. Harris and EuroRotaNet, personal communication).
The differences in incidence of rotavirus and norovirus seen in our study compared with previously published studies that used comparable detection methods is explained by the year on year variations characteristic of enteric virus infections. However, there are also differences in the design of the structured surveillance studies. Whereas, the present study was conducted throughout an entire year, the study in East Anglia was conducted in three consecutive winter seasons (December–May). Analysis of the samples obtained between December and May of the present study showed incidences of 29·7% and 17·8% for rotavirus and norovirus respectively (data not shown), providing more comparable data. The incidence of norovirus infection reported in the IID study (51·1%) [Reference Amar5] was higher than that reported in a structured surveillance study (13·9%) [Reference Iturriza7] and in the present study (24·5%). The IID study was conducted between 1993 and 1996, at a time when a new virus variant emerged into the human population and was likely to have resulted in an increased incidence of infection similar to that reported in 2002 [Reference Lopman18]. Interestingly, the higher incidence of norovirus infection reported in the present study when compared with the structured surveillance study again may be related to the introduction of a new virus variant in 2006 [Reference Gallimore14, Reference Allen19]. Similar incidences of rotavirus infection were reported for the IID (51·1%) and structured surveillance (47·5%) studies, compared with an incidence of 19% reported in the present study. These results demonstrate the need for surveillance activities to be sustained over several years in order to correctly estimate the true burden of disease associated with enteric pathogens.
In the present study a total of six G-types and 2 P-types were identified in eight different combinations (Fig. 6). If protection through vaccination is associated only with rotavirus proteins that elicit a homotypic neutralizing response then the current monovalent (Rotarix™; GSK, Rixensart, Belgium) and pentavalent (Rotateq™; Merck, PA, USA) vaccines would be expected to provide protection from symptomatic infections with strains containing G1 and P[Reference Djuretic8] and G1, G2, G3, G4 and P[Reference Djuretic8], respectively. If protection is in part associated with non-neutralizing antibodies such as those to VP6 and/or NSP4 proteins, then the monovalent and pentavalent vaccines may also protect against disease associated with the majority of common strains co-circulating in the human population, but only the pentavalent vaccine may offer protection from disease associated with strains of subgroup 1/NSP4 genotype A. Four of the G types and one of the P types found in the present study are included in the pentavalent vaccine whereas the monovalent vaccine comprises G1 and P[Reference Djuretic8] which represented 65·8% of the strains detected in the study.
Although all the specimens in our study were submitted from symptomatic children and in the absence of any other pathogen a presumption of cause and effect is reasonable, asymptomatic infections with enteric pathogens were not uncommon [Reference de Wit1, Reference de Wit2, Reference Amar5, Reference de Wit20]. The agent associated with symptoms in multiple infections and the identification of asymptomatic infections may have to rely on the development of robust quantitative methods and the determination of a clinically relevant pathogen concentration.
IID in children is common. The average weekly incidence of IID reported in the WRS in children aged <5 years between 2001 and 2007 was 117·6/100 000. Extrapolated to the population of England & Wales this is equivalent to 186 755 episodes of IID per year. Mean weekly incidence in each 4-week period has fallen gradually since 2001 (Fig. 2). Over the period 1996–2004 there were about 18 000 hospital episodes for IID (~10% of GP episodes) and death certificate data (1996–2004) averaging 15 deaths per annum. Previously, the number of hospitalizations and deaths due to rotavirus infections in the United Kingdom were estimated at 12 787 and 14, respectively in one study [Reference Soriano-Gabarro21], and 13 697 and between 3·2 and 3·5 depending on the method used, respectively, in two further studies [Reference Harris22, Reference Jit23].
The declining incidence in recent years may be largely due to improved standards of hygiene and the use of disposable nappies, but changes in family size may also be relevant. Over the periods 1993–1996 [Reference Amar5] and 2006–2007 (present study) the incidence of infection with viruses in children aged <5 years declined from 68·8% to 63·1% and bacterial and parasitic infections fell from 25·7% to 2·5% and 5·5% to 0·2%, respectively. Moreover, changes to the structure of health services such as the introduction of NHS Direct and the reorganization of the out-of-hours medical provision may have contributed to a fall in the number of IID cases reported through the WRS system. The burden of disease is, however, not solely based on the numbers attending GPs or admitted to hospital but includes the many children managed without recourse to medical care. The non-direct costs attributable to absence from work and the distress of the individuals concerned are also a significant part of the burden [Reference Lorgelly24].
We estimate that from 2001 to 2007 there were on average 186 755 new cases of IID diagnosed per year by GPs across England and Wales. Using the virological data collected from this and the IID study [Reference Amar5], between 19% and 51·1% of these cases would be attributable to rotavirus depending on the year of study. Data from the vaccine trials reported an efficacy of 86·0% against clinic visits for rotavirus gastroenteritis and 83·8% against rotavirus-related medical attention for Rotateq and Rotarix, respectively [Reference Vesikari25, Reference Vesikari26]. Therefore, the introduction of a rotavirus vaccine in England and Wales could result in a reduction of between 29 735 and 82 071 cases of IID in young children diagnosed in primary care each year. In addition, the reported vaccine efficacy against hospital admissions due to rotavirus infection was of 94·7% and 96% for Rotateq and Rotarix, respectively [Reference Vesikari25, Reference Vesikari26]. In addition, vaccination would prevent between 12 109 and 13 149 hospital admissions, according to the estimates of Soriano-Gabarro et al. [Reference Soriano-Gabarro21] or Harris et al. [Reference Harris22].
ACKNOWLEDGEMENTS
The Nappy Study was funded through an unrestricted grant from Sanofi Pasteur MSD. We thank the GPs and practice staff who participated in the study, and Paul Laidler of the HPA Virus Reference Division. We thank Dr Nicholas Kitchin (Sanofi Pasteur MSD) for support in attending project steering group meetings.
DECLARATION OF INTEREST
D. F. has received fees from Sanofi Pasteur MSD for unrelated services. A. E., M. I. G. and J. G. have received financial assistance to attend related symposia.











