Introduction
Globally and locally, achieving the control of historically impactful infectious diseases of livestock continues to frustrate producers and challenge animal health specialists. A core requirement of a successful control program is the ongoing collection of disease data from populations. Schwabe (Reference Schwabe1982) describes this as the process of establishing baseline levels ‘against which effects of intervention (control) efforts can be measured’.
The ongoing burden of disease in endemic areas and the expansion of infectious agents into previously free areas expose the frailty of current surveillance and response/control programs (Backer et al., Reference Backer, Hagenaars, van Roermund and de Jong2009; Lee, Reference Lee2015; Saeed et al., Reference Saeed, Kanwal, Arshad, Ali, Shaikh and Abubaka2015; Neira et al., Reference Neira, Brito, Mena, Culhane, Apel, Max, Perez, Moreno, Mathieu, Johow, Badia, Torremorell, Medina and Ortega2017). Foot-and-mouth disease virus (FMDV) was identified in 1897, but 116 years later, endemic FMDV losses were estimated at $6.5–$21 billion dollars annually and only 66 of the 181 (36.5%) OIE-member countries are ‘FMD free where vaccination is not practiced’ (Longjam et al., Reference Longjam, Deb, Sarmah, Tayo, Awachat and Saxena2011; Knight-Jones and Rushton, Reference Knight-Jones and Rushton2013; OIE, 2017a). Classical swine fever virus (CSFV) was identified in 1903 (de Schweinitz, Dorset, Reference de Schweinitz and Dorset1903), but in 2017, just 32 of the 181 (17.7%) OIE-member countries are considered free of CSFV (OIE, 2017b). This, despite the profound global economic burden of CSFV and the clear benefits of eradication, e.g. the benefit:cost ratio of CSFV eradication in the USA was estimated at ≥13.2 (USDA, 1981; Pinto et al., Reference Pinto, Depner and Vargas-Teran2011). Initially identified on the basis of outbreaks of unknown origin in the 1980s, porcine reproductive and respiratory syndrome virus (PRRSV) was isolated in 1991 and has become endemic in most major pork-producing regions of the world (Wensvoort et al., Reference Wensvoort, Terpstra, Pol, ter Laak, Bloemraad, de Kluyver, Kragten, van Buiten, den Besten, Wagenaar, Broekhuijsen, Moonen, Zetstra, de Boer, Tibben, de Jong, van ‘t Veld, Greenland, van Gennep, Voets, Verheijden and Braamskamp1991; Zimmerman et al., Reference Zimmerman, Benfield, Dee, Murtaugh, Stadejek, Stevenson, Torremorell, Zimmerman, Karriker, Ramirez, Schwartz and Stevenson2012). Holtkamp et al. (Reference Holtkamp, Kliebenstein, Neumann, Zimmerman, Rotto, Yoder, Wang, Yeske, Mowrer and Haley2013) estimated the US pork producers’ losses to PRRSV at $664 million annually. Nathues et al. (Reference Nathues, Alarcon, Rushton, Jolie, Fiebig, Jimenez, Geurts and Nathues2017) estimated losses to European producers at €126.79 per sow per year and €3.77 per pig marketed in herds with ‘slight’ PRRS.
A promising trend in the evolution toward more efficient and effective livestock disease surveillance is the increased use of aggregate samples (Thurmond and Perez, Reference Thurmond and Perez2006; Strutzberg-Minder et al., Reference Strutzberg-Minder, Boehmer, Fischer, Homuth, Gomez-Duren, Finger and Genzow2015; Gibert et al., Reference Gibert, Martin-Valls and Mateu2017; Rotolo, et al., Reference Rotolo, Sun, Wang, Gimenez-Lirola, Baum, Gauger, Harmon, Hoogland, Main and Zimmerman2017). By definition, an aggregate sample represents two or more animals at a specific location and time, e.g. bulk tank milk and pen-based oral fluid samples. As opposed to individual animal samples, e.g. probang samples, swabs, or blood samples, aggregate samples can be collected without animal restraint. The use of aggregate samples in veterinary surveillance has grown in tandem with developments in diagnostic technology, e.g. nucleic acid-based assays and antibody assays specifically adapted to these specimens. The purpose of this article is to review the use of bulk tank milk and pen-based oral fluids in infectious disease surveillance of livestock populations.
Bulk tank milk samples
Bulk tanks are designed to cool, agitate, and store milk in bovine, ovine, and caprine grade A dairies. Among other requirements of the Pasteurized Milk Ordinance (U.S. Food and Drug Administration, 2015), bulk tanks must chill milk (4.4–7°C) within 2 h of collection and maintain this range thereafter. The size and number of bulk tanks vary among farms as a function of the number of animals in the herd or flock, but larger operations may have multiple tanks capable of storing thousands of gallons of milk. Milk haulers may collect once a day, more than once a day, or every other day, depending on the farm's storage capacity and milk production levels. Regardless of the collection schedule, bulk tanks must be emptied, cleaned, and sanitized at least every 72 h (Bickett-Weddle et al., Reference Bickett-Weddle, Keplinger and Sanchez2011; U.S. Food and Drug Administration, 2015).
In the context of disease surveillance, samples from bulk milk tanks represent the lactating cows in the herd (Sekiya et al., Reference Sekiya, Zintl and Doherty2013). Depending on the governmental standards or ordinances, tanks are agitated for ≥10 min after which samples are collected aseptically from the top of the tank using a sterile pipette, syringe, or sanitized dipper (Bickett-Weddle et al., Reference Bickett-Weddle, Keplinger and Sanchez2011; U.S. Food and Drug Administration, 2015). Although bulk tank milk samples do not represent dry cows or cows on milk withhold, they provide an economical, convenient, and timely approach for the detection of specific pathogens and/or estimation of herd prevalence (Olde Riekerink et al., Reference Olde Riekerink, Barkema, Veenstra, Poole, Dingwell and Keefe2006; Sekiya et al., Reference Sekiya, Zintl and Doherty2013; Lanyon et al., Reference Lanyon, McCoy, Bergman and Reichel2014; Collins et al., Reference Collins, Grant, Barrett, Doherty, Hallinan and Mee2017). Economically significant pathogens detectable in bulk tank milk samples and reported in the refereed literature are discussed below and listed in Table 1.
Table 1. Pathogens detected in bulk tank milk
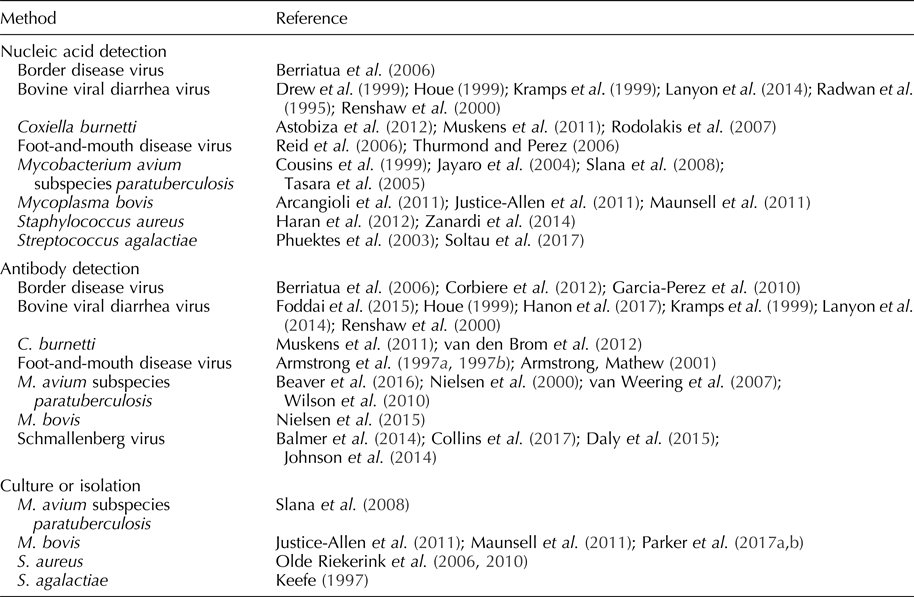
Schmallenberg virus
Schmallenberg virus (SBV) is an arthropod vector-borne orthobunyavirus first detected in dairy herds in Germany and the Netherlands in 2011 (Balmer et al., Reference Balmer, Vogtlin, Thur, Buchi, Abril, Houmard, Danuser and Schwermer2014; Gubbins et al., Reference Gubbins, Turner, Baylis, van der Stede, van Schaik, Cortinas Abrahantes and Wilson2014; Johnson et al., Reference Johnson, Bradshaw, Boland and Ross2014; Daly et al., Reference Daly, King, Tarlinton, Gough, Maddison and Blowey2015). SBV infection causes abortions, congenital malformations, diarrhea, and fever in bovine, ovine, and caprine species (Johnson et al., Reference Johnson, Bradshaw, Boland and Ross2014; Daly et al., Reference Daly, King, Tarlinton, Gough, Maddison and Blowey2015; Collins et al., Reference Collins, Grant, Barrett, Doherty, Hallinan and Mee2017). The duration of SBV viremia is relatively short, i.e. an average of 3–4 days (Gubbins et al., Reference Gubbins, Turner, Baylis, van der Stede, van Schaik, Cortinas Abrahantes and Wilson2014), but SBV serum-neutralizing antibodies can be detected in cattle for as long as 24 months post-infection (Elbers et al., Reference Elbers, Stockhofe-Zurwieden and van der Poel2014). The detection of SBV nucleic acid has not been reported in milk, but antibodies to SBV can be detected in an individual cow and bulk tank milk samples using commercial indirect enzyme-linked immunosorbent assay (ELISAs) (Balmer et al., Reference Balmer, Vogtlin, Thur, Buchi, Abril, Houmard, Danuser and Schwermer2014; Johnson et al., Reference Johnson, Bradshaw, Boland and Ross2014; Daly et al., Reference Daly, King, Tarlinton, Gough, Maddison and Blowey2015). Although test performance estimates are not available (diagnostic sensitivity, diagnostic specificity), results of bulk tank milk ELISA testing were predictive of within-herd seroprevalence and herd immunity (Collins et al., Reference Collins, Grant, Barrett, Doherty, Hallinan and Mee2017). Analyses based on bulk tank milk testing results have been used to assess the spatial distribution, rate of spread, direction of the spread, and effect of farm altitude on the prevalence of SBV (Balmer et al., Reference Balmer, Vogtlin, Thur, Buchi, Abril, Houmard, Danuser and Schwermer2014; Johnson et al., Reference Johnson, Bradshaw, Boland and Ross2014).
Bovine viral diarrhea virus
First described in the 1940s, bovine viral diarrhea virus (BVDV) is a pestivirus transmitted through direct contact or fetal (in utero) infection (Goens, Reference Goens2002). Clinical signs of BVDV include watery and/or bloody diarrhea, dehydration, pyrexia, tenesmus, tachypnea, and ulcers of the muzzle, lips, oral cavity, and/or nares (Goens, Reference Goens2002).
BVDV antibodies can be detected in bulk tank milk samples using blocking, indirect, or competitive ELISAs (Houe, Reference Houe1999; Kramps et al., Reference Kramps, van Maanen, van de Wetering, Stienstra, Quak, Brinkhof, Rùnsholt and Nylin1999; Renshaw et al., Reference Renshaw, Ray and Dubovi2000; Lanyon et al., Reference Lanyon, McCoy, Bergman and Reichel2014; Foddai et al., Reference Foddai, Enøe, Stockmarr, Krogh and Uttenthal2015; Hanon et al., Reference Hanon, De Baere, De la Ferte, Roelandt, Van der Stede and Cay2017). A Danish blocking ELISA demonstrated a diagnostic sensitivity of 100% and diagnostic specificity of 62% when testing bulk tank milk samples from herds with a BVDV prevalence of 26% (Foddai et al., Reference Foddai, Enøe, Stockmarr, Krogh and Uttenthal2015). Diagnostic sensitivities and specificities of competitive ELISAs were reported as 97–100% and 99%, respectively; whereas the diagnostic sensitivities and specificities of indirect ELISAs were reported as 94–100% and 98% (Hanon et al., Reference Hanon, De Baere, De la Ferte, Roelandt, Van der Stede and Cay2017). As with SBV, bulk tank milk ELISA results were highly associated with herd seroprevalence (Lanyon et al., Reference Lanyon, McCoy, Bergman and Reichel2014).
Persistently infected (PI) animals, the result of fetal infection during the first trimester of pregnancy (immunotolerance), serve as a continuous source of infection (Houe, Reference Houe1999; Fray et al., Reference Fray, Paton and Alenius2000; Renshaw et al., Reference Renshaw, Ray and Dubovi2000). PI cows produce little-to-no BVD antibody, but continuously shed real-time reverse transcription polymerase chain reaction (RT-rtPCR)-detectable levels of BVDV in milk (Radwan et al., Reference Radwan, Brock, Hogan and Smith1995; Kramps et al., Reference Kramps, van Maanen, van de Wetering, Stienstra, Quak, Brinkhof, Rùnsholt and Nylin1999; Houe, Reference Houe1999; Renshaw et al., Reference Renshaw, Ray and Dubovi2000). Drew et al. (Reference Drew, Yapp and Paton1999) reported 100% diagnostic sensitivity and specificity for PCR-based detection of BVDV RNA in bulk tank milk samples from herds with PI cows.
Strategically, antibody detection is used to identify the herds with circulating BVDV, and nucleic acid detection is used to identify the herds with PI cattle (Lanyon et al., Reference Lanyon, McCoy, Bergman and Reichel2014). Monitoring changes in antibody prevalence has been used to determine whether a BVDV infection is ongoing or recent (Lanyon et al., Reference Lanyon, McCoy, Bergman and Reichel2014). ELISA testing has also been used to monitor declining antibody levels after removal of PI cattle (Houe, Reference Houe1999).
Border disease virus
First reported in England and Wales in 1958 and closely related to BVDV, border disease virus (BDV) is a pestivirus of ovine and caprine species (Nettleton et al., Reference Nettleton, Gilray, Russo and Dlissi1998). BDV is transmitted through direct contact or transplacentally, with infection during early pregnancy resulting in PI offspring (Garcia-Perez et al., Reference Garcia-Perez, Ruiz-Fons, Barandika, Aduriz, Juste and Hurtado2010). Goats are susceptible to BDV, but infection is rare and typically results in abortion (Nettleton et al., Reference Nettleton, Gilray, Russo and Dlissi1998). In sheep, clinical signs of BDV include abortion, stillbirths, and non-viable lambs.
As in the case of BVDV, PI animals shed BDV continuously and do not produce antibodies. Bulk tank milk samples can be tested for BDV by RT-rtPCR; however, estimates of diagnostic performance have not been reported (Berriatua et al., Reference Berriatua, Barandika, Aduriz, Hurtado, Estevez, Atxaerandio and Garcia-Perez2006). Immunocompetent animals produce antibodies detectable in bulk tank milk (Garcia-Perez et al., Reference Garcia-Perez, Ruiz-Fons, Barandika, Aduriz, Juste and Hurtado2010). In one study, the diagnostic sensitivity and specificity of a blocking ELISA for BDV detection in bulk tank milk samples was reported as 100 and 85.2%, respectively (Corbiere et al., Reference Corbiere, Puget, Bernardin, Brugidou and Schelcher2012). A high seroprevalence of BDV in lactating animals suggests the presence of PI animals (Berriatua et al., Reference Berriatua, Barandika, Aduriz, Hurtado, Estevez, Atxaerandio and Garcia-Perez2006). Thus, ELISA testing of bulk tank milk samples provides the means to estimate the prevalence of BDV in flocks and may indirectly reveal the presence of PI animals (Berriatua et al., Reference Berriatua, Barandika, Aduriz, Hurtado, Estevez, Atxaerandio and Garcia-Perez2006; Garcia-Perez et al., Reference Garcia-Perez, Ruiz-Fons, Barandika, Aduriz, Juste and Hurtado2010).
Foot-and-mouth disease virus
FMDV is a highly impactful picornavirus of cloven-hoofed animals (Reid et al., Reference Reid, Parida, King, Hutchings, Shaw, Ferris, Zhang, Hillerton and Paton2006; Thurmond and Perez, Reference Thurmond and Perez2006; Knight-Jones and Rushton, Reference Knight-Jones and Rushton2013). FMDV can be transmitted by direct or indirect contact (Bravo de Rueda et al., Reference Bravo de Rueda, Dekker, Eble and de Jong2014). Clinical signs of FMDV infection include vesicular lesions, decrease in milk yield in lactating cattle, and pyrexia (Armstrong and Mathew, Reference Armstrong and Mathew2001).
FMDV was detected in milk samples from individual cows by RT-rtPCR for 23 days post-inoculation (Reid et al., Reference Reid, Parida, King, Hutchings, Shaw, Ferris, Zhang, Hillerton and Paton2006). Estimates of the diagnostic sensitivity and specificity of RT-rtPCR for the detection of FMDV in bulk tank milk samples has not been reported, but Thurmond and Perez (Reference Thurmond and Perez2006) predicted that RT-rtPCR testing of bulk tank milk samples would detect FMDV 4–7 days earlier than detection based on the recognition/reporting of clinical signs.
FMDV antibodies may be detected in ovine and bovine milk using blocking ELISAs (Armstrong, Reference Armstrong1997a, Reference Armstrong1997b). Estimates for diagnostic sensitivity and specificity of these ELISAs are not available, but Armstrong and Mathew found a statistically significant correlation (r = 0.53) between serum and milk antibody titers (Armstrong and Mathew, Reference Armstrong and Mathew2001). On this basis, these researchers suggested that antibody testing of bulk tank milk samples would be an effective approach for monitoring herd immunity and/or evaluating population susceptibility to FMDV.
Mycobacterium avium subspecies paratuberculosis
Mycobacterium avium subspecies paratuberculosis (MAP) is the etiologic agent of Johne's disease in ruminants (Mortier et al., Reference Mortier, Barkema, Negron, Orsel, Wolf and De Buck2014). Most commonly acquired via fecal–oral transmission, Johne's disease is characterized by enteritis, decreased milk yield, weight loss, diarrhea, and death (Wilson et al., Reference Wilson, Rood, Biswas and Byrem2010; Mortier et al., Reference Mortier, Barkema, Negron, Orsel, Wolf and De Buck2014). A causal role for MAP in Crohn's disease has been postulated, but was neither confirmed nor rejected by an assessment of the available data (Feller et al., Reference Feller, Huwiler, Stephan, Altpeter, Shang, Furrer, Pfyffer, Jemmi, Baumgartner and Egger2007).
MAP is detectable in milk via culture and PCR testing, but culture of bulk tank milk samples is not practical because the procedure is neither diagnostically sensitive nor timely, i.e. culture can take 18–52 weeks (Slana et al., Reference Slana, Kralik, Kralova and Pavlik2008). The most common target of PCR assays is multiple copy insertion sequence IS900 in the MAP genome (Slana et al., Reference Slana, Kralik, Kralova and Pavlik2008). The analytical sensitivity of the IS900 PCR is reported as 5–6 MAP cells ml−1 of bulk tank milk versus 83 MAP cells ml−1 for a PCR targeting F57. However, IS900 PCRs may have issues with analytical specificity because of the homology of this region across mycobacteria species (Cousins et al., Reference Cousins, Whittington, Marsh, Masters, Evans and Kluver1999; Tasara et al., Reference Tasara, Hoelzle and Stephan2005; Slana et al., Reference Slana, Kralik, Kralova and Pavlik2008). Jayaro et al. (Reference Jayaro, Pillai, Wolfgang, Griswold, Rossiter, Tewari, Burns and Hutchinson2004) reported a diagnostic sensitivity of 21% and diagnostic specificity of 50% for bulk tank milk samples using an IS900 PCR. No estimates of diagnostic sensitivity and specificity are available for F57-based PCRs.
ELISA-detectable MAP antibodies are present in bulk tank milk samples, but interpretation of testing results has not been clearly established (Nielsen et al., Reference Nielsen, Thamsborg, Houe and Bitsch2000; van Weering et al., Reference van Weering, van Schaik, van der Meulen, Waal, Franken and van Maanen2007; Wilson et al., Reference Wilson, Rood, Biswas and Byrem2010; Beaver et al., Reference Beaver, Cazer, Ruegg, Grohn and Schukken2016). Regardless, some researchers believe that ELISA testing of bulk tank milk samples can be used effectively by monitoring changes over time (van Weering et al., Reference van Weering, van Schaik, van der Meulen, Waal, Franken and van Maanen2007; Beaver et al., Reference Beaver, Cazer, Ruegg, Grohn and Schukken2016). Alternatively, Beaver et al. (Reference Beaver, Cazer, Ruegg, Grohn and Schukken2016), suggested the concurrent use of both assays for bulk tank milk monitoring programs for MAP (Beaver et al., Reference Beaver, Cazer, Ruegg, Grohn and Schukken2016). Thus, herds with positive PCR results and high ELISA titers reflected active infection; whereas, herds with positive PCR results but low ELISA titers reflected environmental contamination (Beaver et al., Reference Beaver, Cazer, Ruegg, Grohn and Schukken2016).
Coxiella burnetii (Q fever)
Coxiella burnetii is an obligate, intracellular rickettsial organism and the cause of Q fever in animals and humans (Kim et al., Reference Kim, Kim, Lafferty and Dubovi2005). Infection with C. burnetii results in reproductive disease, including metritis and infertility in cattle and abortion in goats and sheep (Kim et al., Reference Kim, Kim, Lafferty and Dubovi2005; Rodolakis et al., Reference Rodolakis, Berri, Hechard, Caudron, Souriau, Bodier, Blanchard, Camuset, Devillechaise, Natorp, Vadet and Arricau-Bouvery2007). Shedding patterns of C. burnetii in milk is species-dependent and varies among cattle, sheep, and goats (Rodolakis et al., Reference Rodolakis, Berri, Hechard, Caudron, Souriau, Bodier, Blanchard, Camuset, Devillechaise, Natorp, Vadet and Arricau-Bouvery2007). Cattle shed C. burnetii in milk for several months, goats shed for a shorter time, and sheep do not reliably shed in milk (Rodolakis et al., Reference Rodolakis, Berri, Hechard, Caudron, Souriau, Bodier, Blanchard, Camuset, Devillechaise, Natorp, Vadet and Arricau-Bouvery2007; Astobiza et al., Reference Astobiza, Ruiz-Fons, Pinero, Barandika, Hurtado and Garcia-Perez2012). Antibody to and nucleic acids of C. burnetii are detectable in bulk tank milk samples with ELISA and PCR, respectively (Rodolakis et al., Reference Rodolakis, Berri, Hechard, Caudron, Souriau, Bodier, Blanchard, Camuset, Devillechaise, Natorp, Vadet and Arricau-Bouvery2007; van den Brom et al., Reference van den Brom, van Engelen, Luttikholt, Moll, van Maanen and Vellema2012). Muskens et al. (Reference Muskens, van Engelen, van Maanen, Bartels and Lam2011) reported diagnostic sensitivity and specificity of 82 and 70%, respectively, when testing bulk tank milk samples by a commercial real-time PCR. The diagnostic sensitivity and specificity of a commercial C. burnetii antibody ELISA for bulk tank milk was reported as 88.2 and 94.6%, respectively, using manufacturer-recommended cutoffs (van den Brom et al., Reference van den Brom, van Engelen, Luttikholt, Moll, van Maanen and Vellema2012). When used in combination, ELISA testing of bulk tank milk samples can be used to determine herd exposure and estimate prevalence of C. burnetii, while PCR testing can be used to determine shedding and prevalence (Muskens et al., Reference Muskens, van Engelen, van Maanen, Bartels and Lam2011; Astobiza et al., Reference Astobiza, Ruiz-Fons, Pinero, Barandika, Hurtado and Garcia-Perez2012).
Detection of bacterial pathogens associated with mastitis
Streptococcus agalactiae is a highly contagious, obligate pathogen of the bovine mammary gland and a cause of subclinical and clinical mastitis (Keefe, Reference Keefe1997; Phuektes et al., Reference Phuektes, Browning, Anderson and Mansell2003; Olde Riekerink et al., Reference Olde Riekerink, Barkema, Veenstra, Poole, Dingwell and Keefe2006; Mweu et al., Reference Mweu, Nielsen, Halasa and Toft2012). Streptococcus agalactiae may be detected in bulk tank milk samples by culture or PCR (Keefe, Reference Keefe1997; Phuektes et al., Reference Phuektes, Browning, Anderson and Mansell2003). As reviewed by Phuektes et al. (Reference Phuektes, Browning, Anderson and Mansell2003), estimates of the diagnostic sensitivity of culture range from 20 to 84%. Estimates of the diagnostic sensitivity and specificity are not available, but as would be expected, testing multiple bulk tank milk samples was shown to increase the likelihood of detecting S. agalactiae by PCR (Phuektes et al., Reference Phuektes, Browning, Anderson and Mansell2003; Soltau et al., Reference Soltau, Einax, Klengel, Katholm, Failing, Wehrend and Donat2017). ELISA-detectable S. agalactiae antibodies have been reported in individual milk samples, but this approach has not been evaluated for bulk tank milk testing (Logan et al., Reference Logan, Meneely and Mackie1982).
Staphylococcus aureus is an opportunistic pathogen and a cause of subclinical and clinical mastitis in cattle, sheep, and goats (Olde Riekerink et al., Reference Olde Riekerink, Barkema, Veenstra, Poole, Dingwell and Keefe2006; Haran et al., Reference Haran, Godden, Boxrud, Jawahir, Bender and Sreevatsan2012; Zanardi et al., Reference Zanardi, Caminiti, Delle Donne, Moroni, Santi, Galletti, Tamba, Bolzoni and Bertocchi2014; Merz et al., Reference Merz, Stephan and Johler2016). As reviewed by Olde Riekerink et al. (Reference Olde Riekerink, Barkema, Scholl, Poole and Kelton2010), culture of bulk tank milk for S. aureus had an estimated diagnostic sensitivity of 21–42% and a diagnostic specificity of 100%. Repeated sampling is recognized to improve the probability of detection by culture (Olde Riekerink et al., Reference Olde Riekerink, Barkema, Veenstra, Poole, Dingwell and Keefe2006, Reference Olde Riekerink, Barkema, Scholl, Poole and Kelton2010). PCR testing of bulk tank milk samples can be used to detect S. aureus, estimate herd prevalence of the infection, and assess for the presence of methicillin-resistant strains (Haran et al., Reference Haran, Godden, Boxrud, Jawahir, Bender and Sreevatsan2012). The diagnostic sensitivity and specificity of PCR testing for S. aureus in bulk tank milk samples is reported at 99 and 67%, respectively (Zanardi et al., Reference Zanardi, Caminiti, Delle Donne, Moroni, Santi, Galletti, Tamba, Bolzoni and Bertocchi2014). Using individual milk, ELISA testing for antibodies against S. aureus may be used to as a screening tool to detect infected animals (Fox and Adams, Reference Fox and Adams2000).
Mycoplasma bovis is a highly pathogenic mycoplasma causing both mastitis and respiratory disease in adult cattle (Parker et al., Reference Parker, House, Hazelton, Bosward and Sheehy2017a). Mycoplasma bovis is detectable in bulk tank milk samples by culture, but the assay can take 7–10 days and overgrowth of bacteria is problematic (Parker et al., Reference Parker, House, Hazelton, Bosward and Sheehy2017a, Reference Parker, House, Hazelton, Bosward, Morton and Sheehy2017b). The diagnostic sensitivity of M. bovis culture is reported as 50%, with diagnostic specificity estimates as high as 100% (Justice-Allen et al., Reference Justice-Allen, Trujillo, Goodell and Wilson2011; Maunsell et al., Reference Maunsell, Woolums, Francoz, Rosenbusch, Step, Wilson and Janzen2011). The diagnostic sensitivity and specificity of M. bovis PCR for individual milk samples is reportedly 100 and 99.3%, respectively, but estimates of PCR performance for bulk tank milk samples have not been reported (Cai et al., Reference Cai, Bell-Rodgers, Parker and Prescott2005). PCR testing allows for more rapid detection of M. bovis versus culture and herd prevalence estimates can be extrapolated from the results (Arcangioli et al., Reference Arcangioli, Chazel, Sellal, Botrel, Bezille, Poumarat, Calavas and Le Grand2011). A commercial antibody ELISA is available for bulk tank milk testing, and estimates for diagnostic sensitivity and specificity are 60.4 and 97.3%, respectively (Nielsen et al., Reference Nielsen, Petersen, Nielsen, Halasa and Toft2015). The combination of PCR and ELISA testing can reveal M. bovis infection in a herd and is an effective approach for surveillance (Nielsen et al., Reference Nielsen, Petersen, Nielsen, Halasa and Toft2015).
Oral fluid samples
Oral fluids are collected from swine or cattle by providing access to a rope suspended in the pen, then recovering the sample for diagnostic testing (Smith et al., Reference Smith, Gray, Moxley, Younts-Dahl, Blackford, Hinkley, Hungerford, Milton and Klopfenstein2004; Prickett et al., Reference Prickett, Kim, Simer, Yoon and Zimmerman2008a, Reference Prickett, Simer, Christopher-Hennings, Yoon, Evans and Zimmerman2008b; Stanford et al., Reference Stanford, Silasi, McAllister and Schwartzkopf-Genswein2009; Prickett et al., Reference Prickett, Cutler, Kinyon, Naberhaus, Stensland, Yoon and Zimmerman2010). Oral fluid samples are an aggregate sample composed of saliva and transudate originating from capillaries within the buccal and gingival mucosa (Prickett et al., Reference Prickett, Kim, Simer, Yoon and Zimmerman2008a). Oral fluids contain both local and serum-derived antibodies and pathogens (Prickett et al., Reference Prickett, Kim, Simer, Yoon and Zimmerman2008a, Reference Prickett, Simer, Christopher-Hennings, Yoon, Evans and Zimmerman2008b; Prickett and Zimmerman, Reference Prickett and Zimmerman2010). In addition, viruses, bacteria, and other test analytes in feed, water, or the environment may be present in oral fluids as a result of normal exploratory behavior (Kittawornrat and Zimmerman, Reference Kittawornrat and Zimmerman2011; Johnson et al., Reference Johnson, Main and Zimmerman2012). This explains the detection of porcine epidemic diarrhea virus (PEDV) in swine oral fluid samples and Escherichia coli and salmonella in cattle (Smith et al., Reference Smith, Moxley, Clowser, Folmer, Hinkley, Erickson and Klopfenstein2005a, Reference Smith, Moxley, Clowser, Folmer, Hinkley, Erickson and Klopfenstein2005b; Renter et al., Reference Renter, Smith, King, Stilborn, Berg, Berezowski and McFall2008; Bjustrom-Kraft et al., Reference Bjustrom-Kraft, Woodard, Gimenez-Lirola, Rotolo, Wang, Sun, Lasley, Zhang, Baum, Gauger, Main and Zimmerman2016). In cattle, oral fluids have been used in the observational studies in feedlot cattle (Renter et al., Reference Renter, Smith, King, Stilborn, Berg, Berezowski and McFall2008; Smith et al., Reference Smith, Moxley, Clowser, Folmer, Hinkley, Erickson and Klopfenstein2005a, Reference Smith, Moxley, Clowser, Folmer, Hinkley, Erickson and Klopfenstein2005b), but have not been routinely utilized in surveillance. In contrast, oral fluids have been used extensively for disease surveillance in swine populations. Therefore, the remainder of this section will focus exclusively on this subject.
Oral fluids can be collected from groups or individual pigs (White et al., Reference White, Rotolo, Olsen, Wang, Prickett, Kittawornrat, Panyasing, Main, Rademacher, Hoogland and Zimmerman2014; Pepin et al., Reference Pepin, Kittawornrat, Liu, Gauger, Harmon, Abate, Main, Carton, Hargrove, Rademacher, Ramirez and Zimmerman2015a, Reference Pepin, Liu, Main, Ramirez and Zimmerman2015b). In group-housed animals, oral fluids offer a higher probability of detection with fewer samples when compared with individual serum samples (Olsen et al., Reference Olsen, Wang, Christopher-Hennings, Doolittle, Harmon, Abate, Kittawornrat, Lizano, Main, Nelson, Otterson, Panyasing, Rademacher, Rauh, Shah and Zimmerman2013). Sampling guidelines for oral fluid collection at the barn or site level have been published (Rotolo et al., Reference Rotolo, Sun, Wang, Gimenez-Lirola, Baum, Gauger, Harmon, Hoogland, Main and Zimmerman2017).
Diagnostic assays optimized for swine oral fluid specimens have been available in North American veterinary diagnostic laboratories since 2010 (Olsen et al., Reference Olsen, Wang, Christopher-Hennings, Doolittle, Harmon, Abate, Kittawornrat, Lizano, Main, Nelson, Otterson, Panyasing, Rademacher, Rauh, Shah and Zimmerman2013; Bjustrom-Kraft et al., Reference Bjustrom-Kraft, Christopher-Hennings, Daly, Main, Torrison and Zimmerman2018). In three North American swine-interest veterinary diagnostic laboratories, the number of oral fluid tests performed increased from 20,963 in 2010 to 369,439 in 2016 (Bjustrom-Kraft et al., Reference Bjustrom-Kraft, Christopher-Hennings, Daly, Main, Torrison and Zimmerman2018). Pathogens detectable in oral fluid samples and reported in the refereed literature are listed in Table 2. Selected pathogens are reviewed below.
Table 2. Pathogens detected in oral fluid
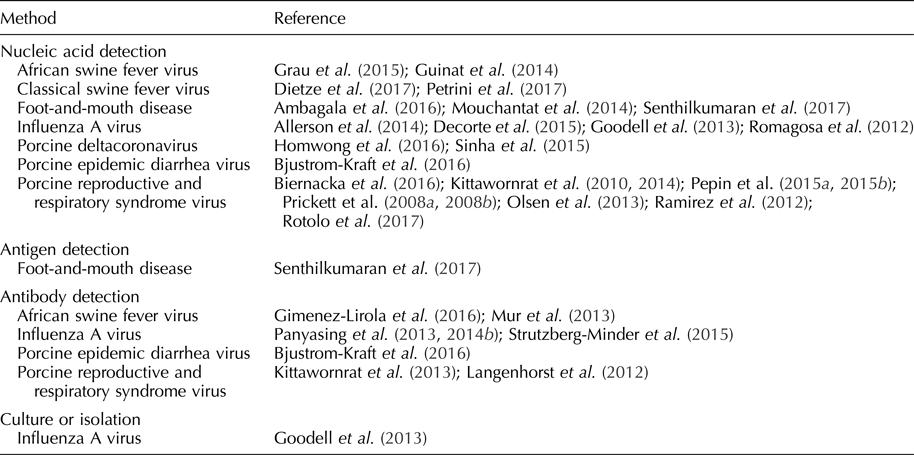
Foot-and-mouth-disease virus
Rapid screening of swine herds is critical in the control of FMDV because pigs aerosolize a large amount of virus compared with cattle and promulgate virus transmission (Stenfeldt et al., Reference Stenfeldt, Diaz-San Segundo, de los Santos, Rodriguez and Arzt2016). Under experimental conditions, FMDV was isolated from swine oral fluids on day post-inoculation (DPI) 1–5 (Senthilkumaran et al., Reference Senthilkumaran, Yang, Bittner, Ambagala, Lung, Zimmerman, Gimenez-Lirola and Nfon2017). By RT-rtPCR, FMDV was detected from one DPI, i.e. prior to the appearance of clinical signs, and up to 21 DPI (Mouchantat et al., Reference Mouchantat, Haas, Bohle, Globig, Lange, Mettenleiter and Depner2014; Senthilkumaran et al., Reference Senthilkumaran, Yang, Bittner, Ambagala, Lung, Zimmerman, Gimenez-Lirola and Nfon2017). RNA was detected in oral fluids one day earlier than oral or nasal swab samples and continued ~7 days longer (Senthilkumaran et al., Reference Senthilkumaran, Yang, Bittner, Ambagala, Lung, Zimmerman, Gimenez-Lirola and Nfon2017). A field-deployable reverse transcription-insulated isothermal PCR has also been used to detect FMDV RNA in oral fluids (Ambagala et al., Reference Ambagala, Fisher, Goolia, Nfon, Furukawa-Stoffer, Ortega Polo and Lung2016). FMDV antigens were detected in oral fluids 1–6 DPI using lateral flow immunochromatographic strip tests and 2–3 DPI using a double-antibody sandwich ELISA (Senthilkumaran et al., Reference Senthilkumaran, Yang, Bittner, Ambagala, Lung, Zimmerman, Gimenez-Lirola and Nfon2017). FMDV IgA was detected in oral fluids using a solid-phase competitive ELISA beginning at 14 DPI (Senthilkumaran et al., Reference Senthilkumaran, Yang, Bittner, Ambagala, Lung, Zimmerman, Gimenez-Lirola and Nfon2017). Pacheco et al. (Reference Pacheco, Butler, Jew, Ferman, Zhu and Golde2010) were not successful in detecting FMDV IgM or IgG in oral fluid samples. Estimates of diagnostic sensitivity and specificity have not been reported for the assays reported in this paragraph. Although FMDV oral fluid assay development is in its early stages, preliminary results support the use of nucleic acid and antibody detection as a method to rapidly screen herds (Ambagala et al., Reference Ambagala, Fisher, Goolia, Nfon, Furukawa-Stoffer, Ortega Polo and Lung2016; Senthilkumaran et al., Reference Senthilkumaran, Yang, Bittner, Ambagala, Lung, Zimmerman, Gimenez-Lirola and Nfon2017).
Classical swine fever virus
CSFV is a pestivirus with significant economic consequences resulting from clinical disease, lost export markets, and costs related to control and eradication efforts (Fernández-Carrión et al., Reference Fernández-Carrión, Ivorra, Martínez-López, Ramos and Sánchez-Vizcaíno2016). CSFV can be transmitted by direct or indirect contact and, depending on the virulence of the strain, causes pyrexia, anorexia, lethargy, conjunctivitis, enlarged and discolored lymph nodes, constipation, and diarrhea in affected pigs (Moennig et al., Reference Moennig, Floegel-Niesmann and Greiser-Wilke2003; Petrini et al., Reference Petrini, Pierini, Giammarioli, Feliziani and De Mia2017). Under experimental settings, CSFV was detected in oral fluids by RT-rtPCR from seven up to 30 DPI, with a higher detection rate in oral fluid than blood samples (40 vs 28%) (Dietze et al., Reference Dietze, Tucakov, Engel, Wirtz, Depner, Globig, Kammerer and Mouchantat2017; Petrini et al., Reference Petrini, Pierini, Giammarioli, Feliziani and De Mia2017). Estimates of diagnostic sensitivity and specificity have not been reported for these assays and, overall, research on CSFV oral fluid diagnostics is in its initial phases.
African swine fever virus
Infection with African swine fever virus (ASFV), the only member of family Asfarviridae, is a cause of fever, hemorrhage, and mortality in domestic and feral pigs (Sanchez-Vizcaino and Neira, Reference Sanchez-Vizcaino, Neira, Zimmerman, Karriker, Ramirez, Schwartz and Stevenson2012; Guinat et al., Reference Guinat, Reis, Netheron, Goatley, Pfeiffer and Dixon2014; Gimenez-Lirola et al., Reference Giménez-Lirola, Mur, Rivera, Mogler, Sun, Lizano, Goodell, Harris, Rowland, Gallardo, Sánchez-Vizcaíno and Zimmerman2016). Transmitted through direct and indirect contact, ASFV is of particular concern because, since its introduction into the Democratic Republic of Georgia in 2007, it has steadily advanced westwardly into Europe via feral swine and threatens to spread eastwardly into China (Guinat et al., Reference Guinat, Reis, Netheron, Goatley, Pfeiffer and Dixon2014; Vergne et al., Reference Vergne, Chen-Fu, Li, Cappelle, Edwards, Martin, Pfeiffer, Fusheng and Roger2017).
Under experimental conditions, ASFV was detected in oral fluid 3–5 DPI by PCR (Guinat et al., Reference Guinat, Reis, Netheron, Goatley, Pfeiffer and Dixon2014; Grau et al., Reference Grau, Schroeder, Mulhern, McIntosh and Bounpheng2015). ASFV antibodies were detected at 11 DPI in individual oral fluid samples by indirect ELISA under experimental conditions (Mur et al., Reference Mur, Gallardo, Soler, Zimmerman, Pelayo, Nieto, Sanchez-Vizcaino and Arias2013). The pattern of antibody response in oral fluids was similar to the pattern seen in serum (Mur et al., Reference Mur, Gallardo, Soler, Zimmerman, Pelayo, Nieto, Sanchez-Vizcaino and Arias2013). ASFV antibodies were also detected using a p30-based indirect ELISA in oral fluids (Gimenez-Lirola et al., Reference Giménez-Lirola, Mur, Rivera, Mogler, Sun, Lizano, Goodell, Harris, Rowland, Gallardo, Sánchez-Vizcaíno and Zimmerman2016). Diagnostic sensitivities and specificities for these assays have not been reported. As in the cases of FMDV and CSFV, further studies are needed to optimize ASFV oral fluid assays and assess their use in the field (Grau et al., Reference Grau, Schroeder, Mulhern, McIntosh and Bounpheng2015).
Porcine reproductive and respiratory syndrome virus
PRRSV is an arterivirus transmitted through direct and indirect contact (Zimmerman et al., Reference Zimmerman, Benfield, Dee, Murtaugh, Stadejek, Stevenson, Torremorell, Zimmerman, Karriker, Ramirez, Schwartz and Stevenson2012). Clinical signs of PRRSV vary based on the age of the pig and the virulence of the isolate. In sows, clinical signs include abortion, stillbirths, anorexia, and mortality (Zimmerman et al., Reference Zimmerman, Benfield, Dee, Murtaugh, Stadejek, Stevenson, Torremorell, Zimmerman, Karriker, Ramirez, Schwartz and Stevenson2012). PRRSV is often an etiological component of the porcine respiratory disease complex in growing pigs (Zimmerman et al., Reference Zimmerman, Benfield, Dee, Murtaugh, Stadejek, Stevenson, Torremorell, Zimmerman, Karriker, Ramirez, Schwartz and Stevenson2012).
The detection of PRRSV nucleic acid in oral fluids has been extensively documented under field and experimental conditions (Prickett et al., Reference Prickett, Kim, Simer, Yoon and Zimmerman2008a, Reference Prickett, Simer, Christopher-Hennings, Yoon, Evans and Zimmerman2008b; Kittawornrat et al., Reference Kittawornrat, Prickett, Chittick, Wang, Engel, Johnson, Patnayak, Schwartz, Whitney, Olsen, Schwartz and Zimmerman2010, Reference Kittawornrat, Panyasing, Goodell, Wang, Gauger, Harmon, Rauh, Desfresne, Levis and Zimmerman2014; Ramirez et al., Reference Ramirez, Wang, Prickett, Pogranichniy, Yoon, Main, Johnson, Rademacher, Hoogland, Hoffmann, Kurtz, Kurtz and Zimmerman2012; Pepin et al., Reference Pepin, Kittawornrat, Liu, Gauger, Harmon, Abate, Main, Carton, Hargrove, Rademacher, Ramirez and Zimmerman2015a, Reference Pepin, Liu, Main, Ramirez and Zimmerman2015b; Rotolo et al., Reference Rotolo, Sun, Wang, Gimenez-Lirola, Baum, Gauger, Harmon, Hoogland, Main and Zimmerman2017). Kittawornrat et al. (Reference Kittawornrat, Prickett, Chittick, Wang, Engel, Johnson, Patnayak, Schwartz, Whitney, Olsen, Schwartz and Zimmerman2010) reported detection in ~10% of experimentally inoculated boars at 24 h post-inoculation by RT-rtPCR. Olsen et al. (Reference Olsen, Wang, Christopher-Hennings, Doolittle, Harmon, Abate, Kittawornrat, Lizano, Main, Nelson, Otterson, Panyasing, Rademacher, Rauh, Shah and Zimmerman2013) evaluated test performance as a function of within-pen prevalence. In pens holding 25 pigs, the probability of detecting PRRSV RNA or PRRSV antibody in pens containing ≥1 positive (4% prevalence) was 62 and 61%, respectively. PRRSV may also be sequenced from oral fluids (Biernacka et al., Reference Biernacka, Karbowiak, Wróbel, Charęza, Czopowicz, Balka, Goodell, Rauh and Stadejek2016).
IgG, IgA, and IgM antibody isotypes were detected in oral fluids collected from individual boars using a commercial PRRS serum antibody indirect ELISA modified for oral fluids (Kittawornrat et al., Reference Kittawornrat, Engle, Panyasing, Olsen, Schwartz, Rice, Lizano, Wang and Zimmerman2013). The pattern of PRRSV antibody ontogeny was similar in serum and oral fluid, with IgM detected in oral fluids at three DPI, IgA at seven DPI, and IgG at eight DPI (Kittawornrat et al., Reference Kittawornrat, Engle, Panyasing, Olsen, Schwartz, Rice, Lizano, Wang and Zimmerman2013). Commercial PRRSV oral fluid ELISAs have since become available. Antibodies were also detected in oral fluid using a fluorescent microsphere immunoassay with a reported diagnostic sensitivity of 92% and diagnostic specificity of 91% (Langenhorst et al., Reference Langenhorst, Lawson, Kittawornrat, Zimmerman, Sun, Li, Christopher-Hennings, Nelson and Fang2012).
Testing of oral fluids can be used to assess the effectiveness of PRRSV control and elimination programs (Biernacka et al., Reference Biernacka, Karbowiak, Wróbel, Charęza, Czopowicz, Balka, Goodell, Rauh and Stadejek2016; Rotolo et al., Reference Rotolo, Sun, Wang, Gimenez-Lirola, Baum, Gauger, Harmon, Hoogland, Main and Zimmerman2017). A distinct advantage of PRRSV oral fluid-based surveillance is that pen-based oral fluid sampling provides a higher probability of detection than individual animal sampling using either RT-rtPCR or ELISA (Olsen et al., Reference Olsen, Wang, Christopher-Hennings, Doolittle, Harmon, Abate, Kittawornrat, Lizano, Main, Nelson, Otterson, Panyasing, Rademacher, Rauh, Shah and Zimmerman2013).
Influenza A virus
Influenza A virus (IAV) is an orthomyxovirus of human beings, horses, sea mammals, birds, and pigs, transmitted via direct and indirect contact (Hughes et al., Reference Hughes, Vincent, Brockmeier, Gauger, Pena, Santos, Braucher, Perez and Loving2015; Neira et al., Reference Neira, Rabinowitz, Rendahl, Paccha, Gibbs and Torremorell2016). IAV in commercial swine herds results in chronic, endemic infection with respiratory or reproductive clinical signs, as well as clinically inapparent infections (Goodell et al., Reference Goodell, Prickett, Kittawornrat, Zhou, Rauh, Nelson, O'Connell, Burrell, Wang, Yoon and Zimmerman2013; Panyasing et al., Reference Panyasing, Goodell, Giménez-Lirola, Kittawornrat, Wang, Schwartz and Zimmerman2013). IAV is an important pathogen to surveil in pigs because of its zoonotic potential (Vincent et al., Reference Vincent, Awada, Brown, Chen, Claes, Dauphin, Donis, Culhane, Hamilton, Lewis, Mumford, Nguyen, Parchariyanon, Pasick, Pavade, Pereda, Peiris, Saito, Swenson, Ven Reeth, Webby, Wong and Ciacci-Zanella2014; Hughes et al., Reference Hughes, Vincent, Brockmeier, Gauger, Pena, Santos, Braucher, Perez and Loving2015).
Under experimental conditions, IAV RNA was detected in swine oral fluids by one DPI and up to 69 DPI (Allerson et al., Reference Allerson, Davies, Gramer and Torremorell2014; Decorte et al., Reference Decorte, Steensels, Lambrecht, Cay and De Regge2015). Decorte et al. (Reference Decorte, Steensels, Lambrecht, Cay and De Regge2015) reported the duration of detection in oral fluids as 14 days longer than detection in nasal swabs by RT-rtPCR (Decorte et al., Reference Decorte, Steensels, Lambrecht, Cay and De Regge2015). Compared with individual nasal swabs, the diagnostic sensitivity and specificity of pen-based oral fluid RT-rtPCR testing was estimated at 80 and 100%, respectively (Romagosa et al., Reference Romagosa, Gramer, Joo and Torremorell2012). Although further optimization is necessary, IAV has also been isolated from oral fluids (Goodell et al., Reference Goodell, Prickett, Kittawornrat, Zhou, Rauh, Nelson, O'Connell, Burrell, Wang, Yoon and Zimmerman2013). Sequencing of IAV from oral fluids has been reported (Panyasing et al., Reference Panyasing, Goodell, Kittawornwat, Wang, Levis, Desfresne, Rauh, Gauger, Zhang, Lin, Azeem, Ghorbani-Nezami, Yoon and Zimmerman2014a). RT-rtPCR testing of oral fluids can be used to track viral circulation and to monitor the effect of vaccination and control programs in commercial swine herds (Goodell et al., Reference Goodell, Prickett, Kittawornrat, Zhou, Rauh, Nelson, O'Connell, Burrell, Wang, Yoon and Zimmerman2013).
Panyasing et al. (Reference Panyasing, Goodell, Giménez-Lirola, Kittawornrat, Wang, Schwartz and Zimmerman2013) reported the ontogeny of IAV IgM, IgA, and IgG in pigs housed under experimental conditions, using isotype-specific indirect ELISAs. Serum and oral fluid IgG responses were highly correlated (r = 0.80) (Panyasing et al., Reference Panyasing, Goodell, Giménez-Lirola, Kittawornrat, Wang, Schwartz and Zimmerman2013). Detection of IAV antibody has also been reported using blocking or competitive ELISA formats (Panyasing et al., Reference Panyasing, Goodell, Wang, Kittawornrat, Prickett, Schwartz, Ballagi, Lizano and Zimmerman2014b; Strutzberg-Minder et al., Reference Strutzberg-Minder, Boehmer, Fischer, Homuth, Gomez-Duren, Finger and Genzow2015). Diagnostic sensitivity and specificity estimates have not been established for these assays. Antibody detection in oral fluids allows for the detection of IAV infection in the absence of clinical signs (Panyasing et al., Reference Panyasing, Goodell, Giménez-Lirola, Kittawornrat, Wang, Schwartz and Zimmerman2013).
Coronaviruses
PEDV is an enteric coronavirus transmitted via the fecal–oral route (Crawford et al., Reference Crawford, Lager, Miller, Opriessnig, Gerber and Hesse2015; Bjustrom-Kraft et al., Reference Bjustrom-Kraft, Woodard, Gimenez-Lirola, Rotolo, Wang, Sun, Lasley, Zhang, Baum, Gauger, Main and Zimmerman2016). Clinical signs of PEDV infection in swine include watery diarrhea, vomiting, and mortality in neonates (Bjustrom-Kraft et al., Reference Bjustrom-Kraft, Woodard, Gimenez-Lirola, Rotolo, Wang, Sun, Lasley, Zhang, Baum, Gauger, Main and Zimmerman2016). In the field, Bjustrom-Kraft et al. (Reference Bjustrom-Kraft, Woodard, Gimenez-Lirola, Rotolo, Wang, Sun, Lasley, Zhang, Baum, Gauger, Main and Zimmerman2016) reported the detection of PEDV nucleic acid in oral fluids from 6 days post-exposure (DPE) to 69 DPE. PEDV was detected 15 days longer in oral fluid samples compared with pen fecal samples, and, compared with individual rectal swabs, oral fluids demonstrated a higher concentration of detectable virus and higher rate of detection. In the same study, Bjustrom-Kraft et al. (Reference Bjustrom-Kraft, Woodard, Gimenez-Lirola, Rotolo, Wang, Sun, Lasley, Zhang, Baum, Gauger, Main and Zimmerman2016) reported the detection of PEDV antibody (IgG and IgA) by 13 DPE in oral fluids. The diagnostic sensitivity and specificity of a PEDV IgG oral fluid ELISA was reported as 69 and 97%, respectively. In contrast, the diagnostic sensitivity and specificity of a PEDV IgA oral fluid ELISA were reported as 100 and 100%, respectively (Bjustrom-Kraft et al., Reference Bjustrom-Kraft, Woodard, Gimenez-Lirola, Rotolo, Wang, Sun, Lasley, Zhang, Baum, Gauger, Main and Zimmerman2016). Although estimates of diagnostic sensitivity and specificity have not been reported, the oral fluid RT-rtPCR is an effective tool to monitor for PEDV presence in herds, and IgA antibody testing offers an effective method to evaluate herd-level immunity (Bjustrom-Kraft et al., Reference Bjustrom-Kraft, Woodard, Gimenez-Lirola, Rotolo, Wang, Sun, Lasley, Zhang, Baum, Gauger, Main and Zimmerman2016).
Like PEDV, porcine deltacoronavirus (PDCoV) is an enteric coronavirus that causes diarrhea and vomiting in pigs (Homwong et al., Reference Homwong, Jarvis, Ching Lam, Diaz, Rovira, Nelson and Marthaler2016). PDCoV can be detected in oral fluids by RT-rtPCR, although estimates of diagnostic sensitivity and specificity are not available (Sinha et al., Reference Sinha, Gauger, Zhang, Yoon and Harmon2015; Homwong et al., Reference Homwong, Jarvis, Ching Lam, Diaz, Rovira, Nelson and Marthaler2016; Zhang, Reference Zhang2016). Homwong et al. (Reference Homwong, Jarvis, Ching Lam, Diaz, Rovira, Nelson and Marthaler2016) reported that the detection of PDCoV nucleic acid in oral fluids was 1.89 times more likely than detection in feces. PDCoV antibody ontogeny in serum and oral fluids has not yet been reported.
Discussion
Globally, the production of livestock – poultry, cattle, and swine – is in the process of shifting from small populations on many farms to large populations on fewer farms (Hoban et al., Reference Hoban, McMillan, Molnar and Parrish1997; Marquer, Reference Marquer2010; Barkema et al., Reference Barkema, von Keyserlingk, Kastelic, Lam, Luby, Roy, LeBlanc, Keefe and Kelton2015; Gale, Reference Gale2017). Readily accessible USDA data from the dairy and swine industries highlight this trend. In 1982, ~275,000 US dairy farms housed ~11,000,000 dairy cows. By 2012, the number of dairy farms dropped to ~64,000, while animal numbers remained relatively stable at ~9,250,000 (USDA, 2014). Pork production has followed the same trend. In 1982, ~330,000 US farms housed ~55,000,000 pigs in 1982. By 2012, the number of farms with pigs declined to ~63,000, while the number of pigs increased to ~66,000,000 (USDA, 2014). Increases in herd size are important to disease control because herd immunity becomes more difficult to achieve as population increases, which in turn leads to pathogen endemicity (LeBlanc et al., Reference LeBlanc, Lissemore, Kelton, Duffield and Leslie2006; Pitzer et al., Reference Pitzer, Aguas, Riley, Loeffen, Wood and Grenfell2016).
Over the same time period, a shift occurred in the movement of livestock across political boundaries. In 1960, 13,500,000 live cattle crossed US state lines for feeding or breeding purposes (Hennessy et al., Reference Hennessy, Roosen and Jensen2005). By 2015, this number had risen to 20,500,000 (USDA, 2017). Similarly, ~2,500,000 pigs were moved across US stateliness in 1960, in contrast to ~52,500,000 moved in 2016 (Shields and Mathews, Reference Shields and Mathews2003; USDA, 2017). Similar patterns have emerged in Europe. For example, Denmark, France, Germany, Italy, the Netherlands, Poland, and Spain cumulatively imported ~910,000 live pigs and exported ~937,000 live pigs in 1961 (FAO, 2017). In contrast, these countries imported ~22,000,000 live pigs and exported ~27,000,000 in 2013 (FAO, 2017). Trends in livestock movement are important because of the well-established role of animal transport in the spread of disease, e.g. the 2001 FMDV outbreak in the UK and, more recently, spread of PEDV throughout the Western Hemisphere (Davies, Reference Davies2015; Guinat et al., Reference Guinat, Relun, Wall, Morris, Dixon and Pfeiffer2016).
The unintended consequences of changes in the structure and management of livestock populations have manifested themselves in the appearance of multifactorial diseases resistant to traditional methods of prevention and control, e.g. bovine and porcine respiratory disease complexes (Schwabe, Reference Schwabe1982; Gardner et al., Reference Gardner, Willeberg and Mousing2002; Hagglund et al., Reference Hagglund, Svensson, Emanuelson, Valarcher and Alenius2006; LeBlanc et al., Reference LeBlanc, Lissemore, Kelton, Duffield and Leslie2006; Bochev, Reference Bochev2007; Edwards, Reference Edwards2010; Pitzer et al., Reference Pitzer, Aguas, Riley, Loeffen, Wood and Grenfell2016). The need to understand complex animal health conditions mandates a shift toward the collection of longitudinal animal health data. New intervention strategies or unanticipated events, e.g. the introduction of an exotic pathogen, can then be evaluated in the context of their impact on baseline values.
Cumulatively, peer-reviewed research supports the conclusion that aggregate samples offer the opportunity to expand the scope of applied surveillance. Testing of bulk tank milk samples provides bovine and small ruminant practitioners and producers the means to monitor disease and estimate herd prevalence and provides animal health researchers the means to evaluate the spatial distribution and rate of disease transmission (Berriatua et al., Reference Berriatua, Barandika, Aduriz, Hurtado, Estevez, Atxaerandio and Garcia-Perez2006; Garcia-Perez et al., Reference Garcia-Perez, Ruiz-Fons, Barandika, Aduriz, Juste and Hurtado2010; Balmer et al., Reference Balmer, Vogtlin, Thur, Buchi, Abril, Houmard, Danuser and Schwermer2014; Johnson et al., Reference Johnson, Bradshaw, Boland and Ross2014; Collins et al., Reference Collins, Grant, Barrett, Doherty, Hallinan and Mee2017). Swine oral fluids offer a more analytically sensitive detection system than individual pig samples, and at a lower cost (Goodell et al., Reference Goodell, Prickett, Kittawornrat, Zhou, Rauh, Nelson, O'Connell, Burrell, Wang, Yoon and Zimmerman2013; Olsen et al., Reference Olsen, Wang, Christopher-Hennings, Doolittle, Harmon, Abate, Kittawornrat, Lizano, Main, Nelson, Otterson, Panyasing, Rademacher, Rauh, Shah and Zimmerman2013). Continued progress toward the goal of effective surveillance using aggregate sampling requires research in two areas: (1) continued development and adaption of diagnostic technology for the most globally impactful diseases of animals and human beings (zoonoses); (2) continued development of statistically valid sampling guidelines including probability of detection estimates by sample size, sampling allocation, and frequency of sampling for farm and regional surveillance.




