Q fever is a zoonotic disease caused by Coxiella burnetii, a Gram-negative obligate intracellular bacterium. It has a worldwide distribution and the main reservoirs of infection are goats, sheep and cattle [Reference Angelakis and Raoult1]. In ruminants, C. burnetii infection can cause a range of reproductive and fertility problems including abortion, with very large numbers of organisms shed at parturition in birth fluids, and fewer numbers shed in milk, faeces and urine [Reference Rodolakis2]. Transmission to humans usually occurs via the respiratory route [Reference Angelakis and Raoult1].
In the Republic of Ireland, there were 13 reported cases of Q fever in humans in 2008 and 17 cases in 2007 [3]. Previous research found 8·5% of Irish Department of Agriculture field and laboratory staff with potential occupational exposure to C. burnetii were seropositive [Reference Reid and Malone4].
Previous research using bulk-milk testing has found prevalences of 21% in England and Wales [Reference Paiba5], and 59% in Denmark [Reference Agger6]. In Northern Ireland, a comprehensive survey of bovine seroprevalence was carried out, with 5182 sera from 273 dairy and beef herds tested by ELISA [Reference McCaughey7]. This survey reported animal- and herd-level prevalences of 6·2% and 48·4%, respectively. Two previous studies of bovine C. burnetii prevalence in the Republic of Ireland were published in 1971, with one reporting 24% seroprevalence from 820 bovine sera [Reference Guven8], and the other finding 0·9% seroprevalence in 2617 bovine sera [Reference Hillary, Shattock and Meenan9]; different animal populations were sampled in these studies, which may explain the discrepancy in results. There is no vaccine for C. burnetii licensed for use in the Republic of Ireland.
The objectives of this study were to estimate the prevalence of exposure to C. burnetii in cattle in the Republic of Ireland, to estimate the true prevalence, to determine whether spatial clustering was present, and to identify potential risk factors associated with exposure.
Two types of sample were collected, blood and milk, using different methods; samples that were collected for other surveillance purposes were subsequently used in this study. In the Republic of Ireland, all female cattle and entire males aged >1 year are blood-sampled annually as part of the national brucellosis surveillance programme. A two-stage randomized sampling system was used to select test sera from these blood samples: 332 herds were randomly selected with geographic stratification from the sampling frame of all herds registered with the Department of Agriculture, Fisheries and Food (DAFF), and five breeding animals aged >1 year were randomly selected from within each of these herds to have their sera forwarded for serological surveillance for bluetongue. These sera were collected in 2008, mostly during the summer. The median age of these animals at sampling was 4 years. There were 18·6% aged <2 years, 15·5% 2 years, 12·6% 3 years, 10·6% 4 years, 9·1% 5 years, 8·5% 6 years, 7·9% 7 years, 4·6% 8 years and 12·5% ⩾9 years; 106 had no date of birth recorded.
Bulk-tank milk samples were collected in November 2009 from dairy herds involved in a herd health project in the Irish Cattle Breeding Federation (ICBF) HerdPlus scheme (www.icbf.com). Stratified random sampling based on geographical location and herd size (based on national Central Statistics Office figures) was used to select 500 farms. Each of these farmers was contacted and asked if they were interested in participating in the herd health project and the 290 who agreed to do so were used in this study. There was no overlap of herds between the blood-sampled and bulk-milk-sampled groups.
The assay for antibodies to C. burnetii was the LSI Q fever ruminant serum/milk ELISA kit (LSI, France), an indirect ELISA which is based on the ovine strain CbO1, rather than the Nine Mile strain (isolated from a tick) used in some other C. burnetii ELISA kits. The manufacturer claims that the use of the CbO1 strain provides superior sensitivity to ELISAs based on the Nine Mile strain (0·87 vs. 0·77) while the specificities are similar (LSI technical file). The assay was performed according to the manufacturer's instructions. A sample/positive control optical density ratio of 0·4 for serum and 0·3 for bulk milk was used as the cut-off for determining positive samples. According to the manufacturer, the sensitivity of the assay for bulk milk is estimated at a prevalence of 10% (LSI technical file), so any herd with >10% of sampled milking cows seropositive for C. burnetii should be detected by bulk-milk testing. The sensitivity of the serum test is reported by the manufacturer as 87% when using serum from confirmed infected animals, while other groups cited the test sensitivity and specificity for serum as 100% and 95%, respectively, compared to PCR [Reference Garcia-Ispierto10, Reference Garcia-Perez11]. An estimate of bulk-milk and serum test specificity was not available from the manufacturer (LSI technical file). A good level of agreement (kappa=0·89) has been reported between serum and milk samples from known infected herds using this test [Reference Guatteo12].
For each animal blood-sampled, the following variables were extracted from the DAFF Animal Health Computer System (AHCS) and Animal Identification and Movement (AIM) computer system: breed, pedigree status, date of birth, gender, number of progeny, still in herd of birth (i.e. homebred) or in different herd (i.e. purchased) at sampling, herd type (dairy, beef), presence of sheep on sampled farm, and number of cattle in sampled herd in 2008. The following variables were extracted from the DAFF AHCS for bulk-milk sample herds: presence of sheep and number of cattle in the herd in January 2010. In addition, the locations of all herds were identified using the DAFF land parcel identification system (LPIS).
Herd size, age, and number of progeny were modelled as continuous variables, and also categorized by quartiles and then modelled as categorical variables. Breed was modelled as several distinct categories, and also categorized as a binary variable as dairy breed (Holstein-Friesian, Holstein-Friesian cross, Jersey cross, Montbeliarde) vs. beef breed (all other breed samples). The choice of continuous or categorical classification was based on comparing univariate models using Akaike's Information Criterion. Presence or absence of sheep on farm, pedigree status, gender, homebred/purchased, and beef or dairy farm type were modelled as binary variables.
Logistic regression analysis was performed on the bulk-milk data using Stata version 10.0 (StataCorp, USA). To account for the within-herd clustering of sera results, a generalized estimating equation logistic regression analysis was used on these data using SAS (SAS Institute, USA). Variables were excluded from the models based on backward selection and likelihood ratio tests (milk data) or generalized score tests (sera) until the model was optimally fitted, with terms dropped from the model if P>0·05. True prevalence estimates were calculated using available estimates of test specificity and sensitivity available elsewhere ([Reference Garcia-Ispierto10, Reference Garcia-Perez11], LSI technical file).
Average nearest neighbour analysis was performed using ArcMap 9.2 (ESRI, USA.).
Table 1 shows the results for bulk milk and serum, with positive results categorized by various characteristics.
Table 1. Prevalence of antibodies to C. burnetii in bulk-milk and serum samples by herd and animal characteristics
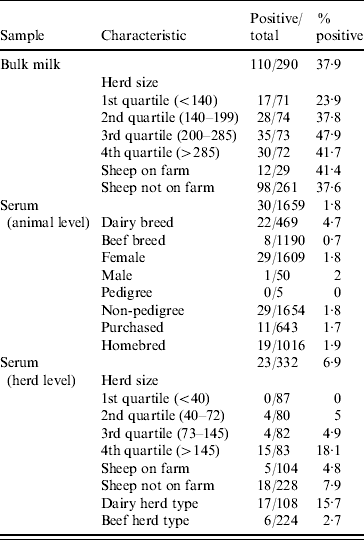
One hundred and ten of the 290 [37·9%, 95% confidence interval (CI) 32·3–43·5] bulk-milk samples contained antibodies to C. burnetii. Data on the specificity of the ELISA for bulk-milk testing were not available, so calculating the true population prevalence from the apparent prevalence is difficult; using the sensitivity value of 0·9 (LSI technical file) and the specificity value of 0·95 given for sera [Reference Garcia-Ispierto10, Reference Garcia-Perez11], the true prevalence is 38·7% (95% CI 33·1–44·3). If a more cautious specificity value of 0·9 is used, the true prevalence is 34·9% (95% CI 29·4–40·4).
The only variable which was statistically significant in the model was herd size categorized by quartile. The odds ratios and confidence intervals are shown in Table 2. The herd size quartiles were 139 and 285, with a median size of 199·5. The presence or absence of sheep on the farm was not significant.
Table 2. Herd-level risk factors for evidence of C. burnetii infection in bulk milk, based on multivariable analysis
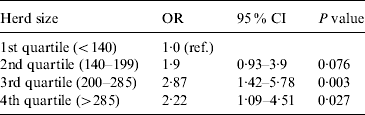
OR, Odds ratio; CI, confidence interval.
Thirty of the 1659 (1·8%) serum samples were positive for C. burnetii antibodies, with 23/332 (6·9%) herds containing at least one positive sample. Five herds contained two positive animals, one herd had three, and the remaining 17 positive herds had one positive animal each. The only variable which was statistically significant in the model was breed type as a binary variable (dairy or beef). The odds ratio and confidence intervals are shown in Table 3. The other variables were not significant, including age, presence of sheep on the farm, pedigree, gender, herd type, herd size, and home-bred vs. purchased.
Table 3. Animal-level risk factors for evidence of C. burnetii infection in serum, based on multivariable analysis

OR, Odds ratio; CI, confidence interval.
Average nearest neighbour analysis indicates that the positive milk and sera samples were distributed randomly throughout the Republic of Ireland.
The recent comprehensive study of Q fever exposure in cattle in Northern Ireland provides a useful comparison to our results [Reference McCaughey7], as agricultural conditions and practices there are quite similar to those in the Republic of Ireland. The Northern Ireland study tested 20 sera per herd (5182 cattle in 273 herds), obtaining a herd prevalence of 48·4% (dairy herd seroprevalence of 64·5%) and animal prevalence of 6·2%. The bulk-milk results presented in our study suggest the dairy herd prevalence in the Republic of Ireland is lower than this, although comparisons between studies using different methodologies are difficult. A recent Danish study [Reference Agger6] of 100 dairy herds found 59% were bulk-milk-positive for C. burnetii antibodies, with 11% intermediate and 30% negative; these authors found no relationship between herd positivity and regional herd density.
The animal prevalence was higher in Northern Ireland (6·2%) than in the Republic of Ireland (1·8%), although the smaller sample size we used (1659 sera) means this must be interpreted with caution. Risk factors for seropositivity in Northern Ireland included increasing age, Friesian breed, large herd size, and being from a dairy herd. We found only dairy breed type (in blood-sampled herds) and increasing herd size (in bulk-milk-sampled herds) were significant risk factors, findings in agreement with those in Northern Ireland. Neither study found an association between the presence of sheep on a farm and exposure to C. burnetii; this may be due to the relatively low numbers of dairy farms with sheep present and the low number of samples per herd. Alternatively, it may suggest low or no exposure to C. burnetii in sheep on sampled farms, or that infection does not readily transmit between cattle and sheep, possibly under the prevailing husbandry conditions in Ireland where segregated calving and lambing occurs. Factors which may have influenced the difference in prevalence between beef and dairy herds include calving conditions, as this is a peak time for transmission, and age profile, which is higher in dairy herds. Further research may be necessary to determine the influence of differing husbandry conditions between the beef and dairy sectors on levels of exposure to C. burnetii. Antibodies to C. burnetii indicate past exposure; acute cases may shed bacteria but have no antibody response, and indeed it has been noted that there is no relationship between bacterial excretion and antibody response [Reference Rodolakis2]. Furthermore, since it was not possible to determine the time of onset of infection, factors which may have influenced infection may subsequently have changed by the time of sampling.
The identification of dairy breed as an animal-level risk factor probably reflects differences in husbandry and management practices between dairy and beef farms, although a possible breed-specific effect cannot be ruled out. Further research is necessary to elucidate the association, which was also identified as a risk factor in Northern Ireland [Reference McCaughey7].
This project was a pilot study designed to provide estimates of prevalence, spatial distribution and related risk factors for bovine C. burnetii infection. Samples were originally collected for bluetongue surveillance, so the sample sizes were not calculated with C. burnetii in mind. The limited sample size undoubtedly reduced the ability to detect significant risk factors.
There was a notable discrepancy between the bulk-milk and serum sample results. Several factors may have influenced this. The selection of five serum samples per herd reduced the herd-level sensitivity for seroprevalence. If the median blood-sampled herd size of 72 and test sensitivity/specificity values of 0·87 and 0·95 are used, herd sensitivity ranges from 0·255 at a herd prevalence of 1% to 0·41 at a herd prevalence of 5%, and herd specificity is 0·745. The low herd sensitivity using serology on five samples may explain much of the discrepancy between dairy herd bulk-milk prevalence (37·9%) and serological herd prevalence (6·9%). Comparing the true prevalences for milk and serum would be helpful, but the low herd specificity estimated above (0·745) means the confidence intervals for estimating true seroprevalence are so wide as to be meaningless. An increase in test specificity would be necessary to overcome this. Another factor, although probably of lesser importance, is the differences in sample selection. Bulk milk can only be taken from dairy herds, whereas only a third of serum samples were from dairy herds. The apparent seroprevalence in dairy herds was 15·7% vs. 2·7% for beef herds. The difference between the dairy herd prevalence as estimated by serum testing and bulk-milk testing was considerable, and the most likely explanatory factor is the lower herd sensitivity when using serum testing. The age profile between the two study populations was different, as only milking cows could contribute to the bulk-milk sample, whereas 18·6% of serum samples (both beef and dairy) were from animals aged <2 years. However, when the sera from these animals aged <2 years were excluded, the serological herd prevalence was recalculated as 7·1%, vs. 6·9% herd prevalence when including all samples. The age difference therefore did not contribute substantially to the discrepancy between the bulk-milk and serum results.
There are two potential sources of selection bias which may affect the results. The dairy farms from which bulk milk was taken were self-selected from a group involved in a farmers' organization. Therefore they may not be fully representative of all Irish dairy herds. The herds which were blood-sampled were only eligible for inclusion in the brucellosis surveillance programme if they had breeding animals aged >1 year; herds composed of steers only or cattle aged <1 year were therefore not included. Nevertheless, the results provide useful and previously unavailable information about bovine Q fever epidemiology in the Republic of Ireland, and contribute to the body of international data on bovine C. burnetii prevalence.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the scientific assistance and advice provided by Dr Michael O'Connor and Goretti McDonagh, the epidemiological advice and useful discussions contributed by Professor Simon More, and the support and advice provided by Dr Paul Collery. Dr Ronan O'Neill and Seamus Power are thanked for kindly providing the serum samples. This project was funded by the Department of Agriculture, Fisheries and Food, Ireland.
DECLARATION OF INTEREST
None.





