INTRODUCTION
Group B streptococcus (GBS), also referred to as Streptococcus agalactiae colonizes the genital and gastrointestinal tract of 10–40% of healthy women. Maternal colonization with GBS is the predominant risk factor for the development of invasive neonatal GBS disease [Reference Schrag1–Reference Heath and Schuchat3].
To date, nine serotypes, based on the GBS capsular polysaccharides (CPS), have been identified (Ia, Ib, II–VIII), and recently a new serotype (IX) has been proposed [Reference Johri4–Reference Slotved6]. In the USA and Europe, serotypes Ia, II, III and V account for 80–90% of all clinical isolates [Reference Lindahl, Stalhammar-Carlemalm and Areschoug2, Reference Brimil7], but serotypes VI and VIII are the most common in Japan [Reference Lachenauer8]. The polysaccharide capsule of GBS isolates is one of the most important virulence factors with anti-phagocytic function [Reference Johri4, Reference Rubens9], hence these polysaccharides are the main components of new multivalent GBS vaccines [Reference Lindahl, Stalhammar-Carlemalm and Areschoug2, Reference Johri4].
The best characterized GBS protein antigens belong to the alpha-like protein (Alp) family. They are named Alpha-C protein, Rib, Alp2, Alp3, Alp4 and Epsilon (Alp1) and are encoded by bca, rib, alp2, alp3, alp4 and epsilon/alp1 genes, respectively [Reference Lindahl, Stalhammar-Carlemalm and Areschoug2, Reference Gherardi5, Reference Persson10, Reference Creti11]. These are a class of surface proteins characterized by internal, long identical tandem repeats [Reference Gherardi5, Reference Creti11] and are encoded by allelic genes presenting a mosaic structure with conserved 5′ and 3′ ends and a highly variable domain generated by intragenic recombination [Reference Gherardi5]. The protein gene profile increases the potential for GBS subtyping [Reference Creti11, Reference Kong12]. The proteins play a role in the pathogenesis of infection and several of these antigens have been proposed as components of GBS conjugate vaccines [Reference Lindahl, Stalhammar-Carlemalm and Areschoug2, Reference Johri4, Reference Heath and Feldman13].
Several molecular-typing methods have been previously described in order to identify the emergence and spread of new GBS clones and to study their genetic population structure. A technique particularly suitable for this purpose is pulsed-field gel electrophoresis (PFGE) of DNA macrorestriction digests [Reference Van Belkum14] and several studies attest to its discrimination and suitability for typing of GBS isolates [Reference Gherardi5, Reference Savoia15, Reference Pillai16].
Knowledge of the epidemiology and molecular characterization of GBS in Poland is limited. Therefore, the main aim of this study was to analyse the genotype distribution of GBS isolates originating from pregnant women in a defined geographical area and to identify CPS and surface protein genes, which might be used in GBS vaccines. Furthermore, the study aimed to determine the correlation of DNA profiles with serotypes and surface protein genes in GBS isolates.
MATERIALS AND METHODS
Study population and specimen collection
The study was conducted from August 2007 to July 2009. Altogether, 1176 women from Southern Poland in their third trimester of pregnancy were screened for GBS colonization according to the ‘swab-based strategy’ recommended by Centers for Disease Control and Prevention (CDC) [Reference Schrag1, Reference Heath and Schuchat3, Reference Kotarski17]. Cultures were obtained by swabbing of both lower vagina and rectum at 35–37 weeks' gestation. Samples were placed in a non-nutrient Amies transport medium (Eurotubo, Spain) and delivered to the microbiology laboratory within 2–4 h. Swabs were first inoculated into a selective broth medium, 3 ml Todd–Hewitt broth (Difco, USA) supplemented with 8 μg/ml gentamicin and 15 μg/ml nalidixic acid (Oxoid, UK), incubated overnight at 37°C, and subcultured onto Columbia 5% sheep blood agar (Difco). Three morphologically similar colonies were selected and tested in the latex-agglutination group kit, Slidex Strepto kit (bioMérieux, France) and API STREP kit (bioMérieux) to identify S. agalactiae. In doubtful cases, identification based on a PCR with species-specific primers was applied [Reference Ke18]. A total of 353 non-consecutive GBS isolates were collected from the vaginal and rectal swabs. When both swabs from an individual were positive for GBS, the vaginal isolate was used for molecular characterization.
Serotyping
Genes encoding CPS Ia, Ib, II–VIII were detected using multiplex PCR with specific primers (Genomed, Poland) according to Poyart et al. [Reference Poyart19].
PCR products were resolved in 2·2% agarose gel with 0·25 μg/ml ethidium bromide (Bio-Rad, USA). The electrophoretic images were processed using QuantityOne (Bio-Rad) software, and analysed with a GelDoc2000 device (Bio-Rad).
Detection of surface protein genes
Surface protein genes were investigated in 169 randomly chosen GBS isolates from vaginal or rectal carriage representing serotypes Ia (n=36), Ib (n=14), II (n=24), III (n=55), IV (n=8) and V (n=32). A multiplex PCR method was used to detect genes bca, rib, epsilon, alp2/3 and alp4 by using five pairs of primers (Genomed), according to Creti et al. [Reference Creti11]. To distinguish between alp2/3 and confirm the presence of alp3 gene the reverse primer Alp3 (5′-TTT TGG TTC GTT GCT ATC CTT AAG C-3′) was used [Reference Gherardi5] with the common forward primer of Creti et al. [Reference Creti11].
PFGE and cluster analysis
One hundred random GBS isolates were typed by PFGE, representing serotypes Ia (n=25), Ib (n=7), II (n=14), III (n=29), IV (n=7) and V (n=18). DNA was digested using SmaI (MBI Fermentas, Latvia) and the fragments separated in a CHEF-DR III device (Bio-Rad), according to the previously described method [Reference Brzychczy-Włoch20]. The genetic similarity between isolates was calculated using Molecular Analyst software (Applied Maths, Belgium), making use of the Jaccard coefficient and unweighted pair-group method with arithmetic mean (UPGMA) as a method for clustering with 3% tolerance and 0·5% optimization settings. Genotype patterns were compared according to the guidelines of van Belkum et al. [Reference Van Belkum14]. Isolates with ⩾90% similarity in PFGE patterns were considered to be indistinguishable and were assigned to the same subtype. Clearly distinguishable PFGE patterns were resolved at a level of 65% similarity on the dendrogram.
Macrolide resistance
The isolates typed by PFGE above were investigated for macrolide resistance phenotype [cMLSB (constitutive), iMLSB (induced) and M] according to Clinical and Laboratory Standards Institute guidelines [21]. The results have been published in a previous study [Reference Brzychczy-Włoch20].
Statistical analysis
For analysis of the frequencies and probability of coincidence, the χ2 test was used. When data were insufficient for the test demands, Fisher's exact test and G 2 (Likelihood ratio test) were used. P values of <0·05 were considered significant. The relative frequency coefficients of PFGE types in the different groups of serotypes and surface protein genes were calculated according to Sokal & Rohlf [Reference Sokal and Rohlf22].
RESULTS
Prevalence of carriage
The GBS carriage rate in the 1176 pregnant women was 30% (353 cases). Simultaneous colonization of the vagina and rectum was demonstrated in 280 women (80%), while vaginal or rectal carriage alone was 33 (9%) and 40 (11%) of cases, respectively.
Serotypes and surface protein genes of colonizing strains
Of the 353 isolates, serotype III was predominant (123, 35%), followed by Ia (71, 20%), V (61, 17%), II (53, 15%), Ib (28, 8%) and IV (17, 5%). Serotypes VI–VIII were not identified. The noted frequencies varied significantly from random distribution (χ2=26·133, P<0·0001). At least one type of a surface protein gene was found in each of the 169 randomly selected strains. The most common gene was epsilon (44, 26%), followed by rib (37, 22%), alp2 (36, 21%), bca (29, 17%) and alp3 (23, 14%); the alp4 gene was not found.
A significant relationship between several surface protein genes and particular serotypes was demonstrated (G 2=249·635, P<0·0001), notably epsilon with Ia, Ib, IV; bca with Ib, II; rib with II, III; alp3 with V; and alp2 with III (Fig. 1).
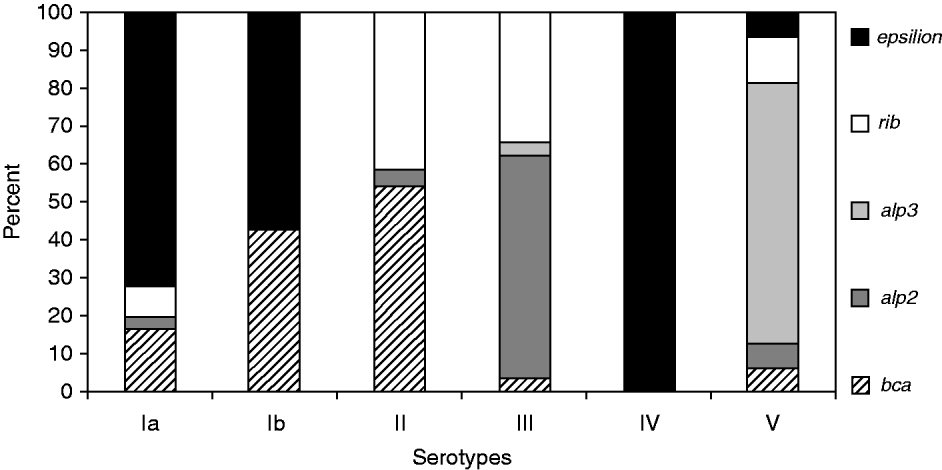
Fig. 1. Diversity of surface protein genes among serotypes in group B streptococci.
Genetic diversity
Figure 2 shows there was high genetic diversity in the selected strains typed by PFGE. At a cut-off level of 90% similarity, 63/100 strains gave uniformly similar DNA profiles and included 21 clusters comprising 2–5 strains with identical profiles; 42 unique profiles were identified within all strains. Analysis of the dendrogram at a level of 65% similarity, revealed 40 different patterns which included 17 clusters of 2–11 strains, and 23 unique patterns (Fig. 2). Of the 17 multiple-strain clusters, nine contained strains of different serotypes. For example, pattern type 1 comprised strains of serotypes V, III and Ia, while pattern type 2 included serotypes Ia and IV, and pattern type 11, serotypes Ib and II.

Fig. 2. Cluster analysis of PFGE patterns following SmaI restriction enzyme digestion of chromosomal DNA from 100 S. agalactiae strains. Strains belonging to the same group (PFGE type) were clustered at a level of 65% similarity (dashed vertical line) and for subtypes at a level of 90% similarity (dotted vertical line).
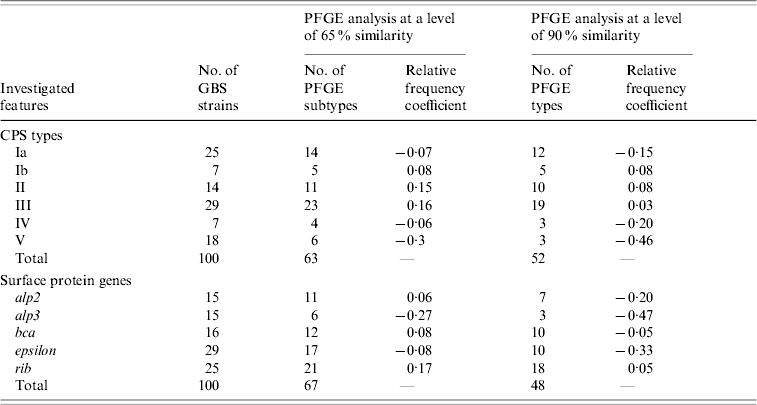
Table 1 shows the relative frequency coefficients of the distribution of PFGE pattern types in serotypes and surface protein genes. A negative value indicates that the genetic diversity of a particular group is less than that of the total strains analysed, while a positive value indicates greater diversity in comparison with the total. Serotypes Ib, II and III were mostly heterogeneous (relative frequency coefficient ⩾0·03), unlike serotype V, especially strains with alp3 gene and macrolide-resistance phenotype, which were mostly genetically homogeneous and appeared to be clonal in genetic profile (relative frequency coefficient –0·46). For example, strains clustering in PFGE pattern types 1 and 4 were mainly of serotype V and represented the majority of macrolide-resistant isolates (Fig. 2).
Table 1. Genetic diversity in 100 GBS strains based on the relative frequency coefficient of PFGE restriction profiles in different groups of capsular polysaccharides (CPS types) and surface protein genes from the Alp family
DISCUSSION
Prophylaxis of neonatal GBS disease is currently based on screening of pregnant women during weeks 35–37 of pregnancy for GBS colonization and then administering the recommended intrapartum antibiotic prophylaxis (IAP) [Reference Schrag1, Reference Heath and Schuchat3]. Contrary to the shortcomings of IAP-based prevention strategies, vaccination has a strong potential for eradicating invasive GBS disease in newborns [Reference Johri4].
In 2008, the Polish Gynaecological Society issued guidelines for prevention of GBS that are generally in accordance with CDC recommendations [Reference Schrag1, Reference Kotarski17]. This paper is the first report after implementation of the Polish Guidelines and the first detailed study of the molecular characterization of GBS isolates in Polish pregnant women.
Our finding of a 30% carriage rate of GBS in 2007–2009 confirms that this colonization is very common in pregnant women and the rate is in accordance with reported rates from other European countries, e.g. between 6·6% in Greece and 36% in Denmark [Reference Tsolia23–Reference Motlova25]. The current rate represents a marked increase over that found in our earlier investigation in Poland, carried out in 2004–2006, where the GBS colonization rate varied from 13% to 17% depending on the detection procedure used [Reference Strus26]. The higher percentage of carriage in the present study is probably due to the use of a more sensitive detection method. Similar results were obtained by Hansen et al. [Reference Hansen24] who demonstrated that the application of a new differential medium in the study of GBS carriage in Danish pregnant women increased the observed carriage rate from 15% to 36% [Reference Hansen24].
Developing a new GBS conjugate vaccine for the prevention of neonatal disease, composed of the most common CPS and surface protein antigens, has been listed as one of the top priorities for vaccine development by the CDC. [Reference Johri4] This approach would also be the most cost-effective strategy and would negate the disadvantages of antibiotics, e.g. growth of antibiotic resistance and allergy in children, and could protect against both early- and late-onset disease in infants [Reference Heath and Schuchat3, Reference Heath and Feldman13]. Because the diversity of CPS and surface protein genes in GBS populations vary by geographical region, ethnic origin and the virulence of clinical isolates [Reference Persson10, Reference Poyart19], epidemiological studies of GBS are important for the formulation of an effective vaccine suitable for use in a range of geographical areas and should therefore be conducted over time in different parts of the world.
In our study serotypes III, V, Ia and II were the most frequent serotypes found, in descending order, accounting for 87% of the total isolates analysed. This distribution corresponds well with other European countries, where these serotypes also account for 80–90% of all clinical isolates, while serotypes VI, VII–IX were seldom observed [Reference Gherardi5, Reference Brimil7]. However, it is worthwhile noting the agreement of our results with data from Slovakia which borders the investigated region in Poland. Slovakia reported a 30% GBS carriage rate in 2004 with a closely similar serotype distribution to that found in our study. Similar results were reported from a survey in Germany in 2006 [Reference Brimil7] although the frequency of serotype III was lower and serotype II higher than found in Poland. Serotypes VI–VIII were not found in the German survey and this suggests the absence of these serotypes in central Europe, unlike Japan, where serotypes VI and VIII are the most common [Reference Lachenauer8]. This feature may be a consequence of different immune responses or diet of the respective populations [Reference Florindo27].
S. agalactiae strains usually exhibit expression of one of the genes from the Alp family, of which proteins Alpha-C, Rib, Alp2, Alp3, Alp4 and Epsilon (Alp1) have been extensively studied [Reference Johri4, Reference Gherardi5, Reference Creti11]. They are characterized by long repetitive elements and mosaicism due to recombination, resulting in sharing of epitopes and antigenic cross-reactivity [Reference Lachenauer28]. This cross-reactivity suggests that a common protein vaccine could provide broad spectrum protection against GBS disease [Reference Seifert29].
In our study, the most common gene from the Alp family was epsilon (27%), followed by rib (21%), alp2 (21%), bca (17%) and alp3 (14%). These results were a little different from data from Norway where the single most common gene identified was rib (42%), followed by alp3 (26%) and epsilon/alp1 (14%). [Reference Persson10] This may reflect geographical variation or the fact that we investigated only strains from carriage and not from infections. The virulence of GBS strains may be associated with surface proteins such as the Alp family, which are significant factors for GBS pathogenicity [Reference Gherardi5, Reference Creti11].
We demonstrated a statistically significant relationship between CPS and several surface protein genes and confirmed previously reported associations between these factors [Reference Creti11, Reference Kong12]. For example, Creti et al. [Reference Creti11] noted a relationship between serotypes Ia, Ib and II with Alpha-C protein as well as serotype III with Rib, and serotypes V and VIII with Alp3, but it was not absolute. Moreover, another study reported associations between epsilon/alp1 gene with serotype Ia; bca with serotypes Ib and II; rib with serotype III, and alp3 with serotype V [Reference Persson10].
A combination of different molecular-typing methods should be considered in order to gain a better understanding of the epidemiology and pathogenesis of GBS isolates [Reference Kong12]. Some studies have used PFGE types as a surrogate for serotyping, presuming that isolates from a single individual with the same PFGE type have the same serotype. However, clonal groups shared by different serotypes has been widely reported [Reference Gherardi5, Reference Pillai16]; this was also noted here as 9/17 multiple strain clusters contained GBS isolates of different serotypes. The capsular serotype of a strain could potentially be altered by as little as the horizontal transfer of a single gene and this polymorphism might not result in a change in PFGE type. Therefore the reliability of using PFGE profiles as predictor of capsular type for isolates from different individuals, even from within the same geographical area, warrants investigation [Reference Pillai16].
Our results confirm the genetic heterogeneity of GBS as shown by other investigators [Reference Gherardi5, Reference Savoia15, Reference Pillai16]. However, the high genetic diversity observed may be explained by the origin of the isolates from asymptomatic carriage in pregnant women. The level of diversity varied with capsular types, with the greatest heterogeneity occurring in types Ib, II and III. Only serotype V, specifically strains with alp3 gene and macrolide-resistance phenotype, were largely homogeneous in genetic profile which is consistent with spread of this clone throughout the community population. These findings are in agreement with the literature [Reference Gherardi5, Reference Pillai16, Reference Florindo27, Reference Diekema30]. Additionally, serotype V, especially strains resistant to erythromycin, has emerged recently as a cause of severe human disease [Reference Gherardi5, Reference Diekema30].
In conclusion, the results obtained have demonstrated that GBS colonization is very common among Polish pregnant women and that GBS isolates from asymptomatic carriage are genetically diverse organisms. The distribution of serotypes and surface protein genes shows considerable similarity to the distribution of GBS isolated in other European countries. Hence, these results may be important as a basis for the formulation of a wide-ranging GBS vaccine.
ACKNOWLEDGEMENTS
This study was supported by a grant from the Polish Ministry of Research and Higher Education no. N N401 042337. The study was approved by Jagiellonian University Bioethical Committee decision no. KBET/143/B/2007. The authors thank the technical staff employed at the Microbiological Diagnostic Laboratory at the Jagiellonian University Medical College for their help in the collection of GBS strains.
DECLARATION OF INTEREST
None.





