INTRODUCTION
Shiga-toxigenic Escherichia coli (STEC) are important zoonotic foodborne pathogens, capable of causing haemorrhagic colitis (HC) and haemolytic uraemic syndrome (HUS). Although, O157:H7 is the major serotype implicated in causing human illness, non-O157 serotypes have also been frequently associated with STEC illness in some geographical regions [Reference Johnson1, 2] with half of all confirmed STEC infections in Europe being caused by non-O157 STEC [3]. Out of 250 non-O157 serogroups, more than 100 are associated with human illness [Reference Eklund4]. Six non-O157 serotypes, i.e. O111:H8 or NM; O103:H2, H11, H25; O26:H11 or NM; O45:H2 or NM; O121:H19 or H7; and O145:NM have been described as being responsible for 71% of non-O157 STEC illnesses in the USA as reported by the CDC [Reference Bosilevac and Koohmaraie5]. A global increase of 60·5% in human disease, caused by non-O157 STEC serotypes was reported between 2000 and 2005 in the Enter-net VTEC database, which included surveillance data from 36 countries, including 31 in Europe as well as from Canada, Australia, New Zealand, Japan and South Africa [6, Reference Coombes7]. STEC have been found to be associated with 75–100% cases of sporadic HUS in Europe, North America, Canada and Latin America [Reference Lopez, Contrini, De Rosa, Kaper and O'Brien8].
Although, cattle are the primary reservoirs of STEC which are isolated from 2·8% to 75% of healthy animals in different farms across USA and Europe [Reference Beutin9–Reference Blanco11], other ruminants such as sheep and goats, also serve as important reservoirs, with isolation rates of 50–60% in a study conducted in Denmark [Reference Beutin9]. STEC are usually transmitted via contaminated foods, mainly undercooked ground beef and unpasteurized dairy products. Other routes of transmission include person-to-person, drinking, and recreational water [Reference Griffin, Blaser, Smith, Ravden, Greenberg and Guerrant12]. Moreover, the dispersion of untreated manure in the environment may be considered a secondary vehicle of transmission of infection to humans [Reference Caprioli13].
A variable repertoire of virulence genes has been reported for both O157 and non-O157 serotypes [Reference Wickham14, Reference Karmali15] out of which stx1, stx2 (Shiga toxins 1 and 2, respectively) and eae (adherence factor intimin) are the most imp ortant [Reference Paton and Paton16].
Multi-locus variable number tandem repeat analysis (MLVA) involves the determination of a number of repeats at multiple loci which are amplified by polymerase chain reaction (PCR). Although pulsed-field gel electrophoresis (PFGE) is considered to be the gold standard technique for molecular typing, MLVA is as a rapid and specific way of differentiating between different isolates of the same serotype. Recently, PulseNet USA established a standardized protocol for MLVA typing of O157:H7 strains [Reference Hyytia-Trees17]. Initially, this method was validated for typing of O157 serotypes alone, as several loci were absent from non-O157 strains. Lindstedt et al. [Reference Lindstedt18] established a generic MLVA assay for all E. coli and Shigella species based on a set of seven variable number tandem repeat (VNTR) loci. These seven loci were used to analyse the E. coli reference collection (ECOR) [Reference Ochman and Selander19]. Bustamante et al. [Reference Bustamante20] have shown the suitability of MLVA for characterizing STEC isolates belonging to several serotypes.
In India, the dynamics of transmission of STEC from animals and the environment are not well understood. There are variable reports on the isolation of STEC from stools of healthy animals (10·5–18%) [Reference Chattopadhyay21, Reference Pal22], diarrhoeic animals (6–37%) [Reference Bandyopadhyay23, Reference Khan24], animal handlers with diarrhoea (3·12%) [Reference Bandyopadhyay23] and raw beef samples (1·8–50%) [Reference Chattopadhyay21, Reference Khan24] from India. A previous study conducted in Kolkata [Reference Khan24] has reported the following serogroups from human isolates: O1, O4, O5, O7, O11, O25, O50, O96, O110, O111, O43, O153, O156 and O159. The only Indian study on the molecular epidemiology of STEC performed by PFGE on 25 isolates concluded that bovine strains are not transmitted to humans [Reference Khan25].
In the current study, which is the first of its type from India, we applied MLVA to gain insight into transmission of non-O157 STEC which were isolated from diverse sources in an epidemiological and ecological survey conducted over 2 years in a large geographical area in North India. To investigate the relationship between major virulence factors and MLVA profiles, the distribution of stx and eae genes was also studied.
MATERIALS AND METHODS
Sample collection and processing
Chandigarh (latitude 30° 43′ N, longitude 76° 47′ E) is an urbanized city but the surrounding areas in the states of Punjab, Haryana and Himachal Pradesh are a mixture of semi-urban and rural population, where animal (cows, buffalos, sheep, goats) rearing is commonly practised. These are major areas of milk and meat production. Samples were collected during November 2007 to October 2009 from an area with a diameter of about 300 km with Chandigarh at its centre. A survey was performed to locate the dairies (Table 1). Purposive sampling was carried out in 59 dairies for animal (cattle, sheep/goat) stool collection. Dairies were classified as small (<25 animals), medium (25–100 animals) and large (>100 animals). Meat samples (chicken, pork, mutton) were collected from local abattoirs/meat retail shops in these areas, as well as from a slaughterhouse under the jurisdiction of the Municipal Corporation Chandigarh, India. Similarly, chutney (ground mixture of raw onion, coriander, mint, garlic, etc.) was collected from street vendors in the same areas. Samples were collected from hilly (⩾670 m above sea level) as well as plane areas (Fig. 1).
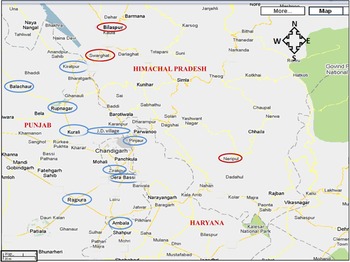
Fig. 1 [colour online]. Sample collection sites: samples were collected from Bilaspur, Swarghat, Kiratpur, Ropar, Kurali, Jayanti Devi, Pinjore, Zirakpur, Derabassi, Naripul, Balachaur, Ambala, Rajpura. Areas circled in red represent hilly areas and areas circled in blue represent plain areas. (The figure has been adapted and modified from https://www.google.com/maps/preview/place/Chandigarh.)
Table 1. Distribution of samples collected and STEC isolated in different geographical areas

STEC, Shiga-toxin Escherichia coli.
* Fifteen out of 128 meat samples collected from Chandigarh slaughterhouse (SH) were positive for STEC. In the slaughterhouse animals are received from the locations listed in column 1.
The total numbers of small dairies in the region were not possible to count since every house has a small dairy. All large dairies were sampled. Hilly areas had small dairies only. Herd size has been characterized as small (<25 animals), medium (25–100 animals) and large (>100 animals).
A total of 650 animal stool (550 cattle, 100 sheep/goat), and 450 meat (mutton, chicken, pork) samples were collected. Forty-five chutney samples were also collected. A total of 600 human stool samples were collected. Stool samples were collected from all patients presenting with bloody diarrhoea and HUS and every fifth case presenting with non-bloody diarrhoea at the Postgraduate Institute of Medical Education and Research.
Processing of samples
Two to three large loopfuls of human or animal stool (cattle, sheep/goat stool) samples were directly inoculated into 3 ml of EC medium (Difco, USA) for enrichment and incubated overnight at 37°C under shaking conditions. Meat samples (10–15 g) were mixed thoroughly with 50 ml EC medium. After 2 h, the broth was transferred into a sterile conical flask and incubated as described above. Human stool samples were also cultured conventionally for other pathogens, e.g. Salmonella, Shigella, Aeromonas and vibrios using standard methods [26].
Screening of enrichment medium by PCR
DNA was extracted from 1 ml of broth culture by the boiling method [Reference Kumar27]. The supernatant was used as a PCR template for the detection of stx1 and stx2 genes [Reference Kumar27]. The cultures positive for either stx1 and/or stx2 by PCR, were serially diluted in 10 mm PBS (pH 7·0), and 100 μl volume of each dilution was spread on sorbitol-MacConkey agar for STEC isolation as described previously [Reference Kumar27]. The STEC isolates were preserved in Luria broth containing 15% glycerol at −70°C.
Serotyping
On the basis of O antigen, isolates were serogrouped by the National Salmonella and Escherichia Centre at the Central Research Institute at Kasauli, India using 16 pools of polyvalent antisera. The culture was then tested with individual monovalent sera. Standard strains (provided by the WHO) were used for raising E. coli sera in rabbits.
MLVA typing
A total of 140 STEC isolates (75 cattle stool, five sheep/goat stool, 39 mutton, 21 human stool strains) were used for MLVA analysis along with reference strain EDL933, obtained from the Pasteur Institute, Paris, France. The seven VNTR loci were amplified according to the protocol described by Lindstedt et al. [Reference Lindstedt18]. After amplification, the products were separated on a 6% denaturing polyacrylamide gel (PAGE) using SequiGen gel apparatus 38 × 50 × 0·4 cm (Bio-Rad Laboratories Inc., USA). The gels were fixed overnight in 10% acetic acid and silver-stained using the Promega silver staining kit (Promega, USA).
MLVA data analysis
Band size of each amplicon was determined by 20 bp DNA marker (Bangalore Genei, India) run along with amplicons in the denaturing PAGE and the allele numbers were assigned for each locus [Reference Lindstedt18]. If no amplification product was detected, the allele was assigned as null allele [Reference Lindstedt18]. To confirm results of the fragment analysis, representative alleles were sequenced using the same primers which were used to amplify the corresponding VNTR regions. The alignment of sequences was obtained using BioEdit v. 7.0.9 (Ibis Biosciences, USA). The tandem repeats for each allele were confirmed using Tandem Repeat Finder software [Reference Benson28]. An allele string was made for each isolate with the order: CVN001-CVN003-CVN004-CVN007-CVN014-CVN015.
The diversity index (DI) based on Nei's marker of diversity was calculated for each locus using the formula DI = 1 – Σ(f)2, where f is the allelic frequency [Reference Noller29]. The discriminatory power of the typing method was evaluated using Simpson's index of diversity (DS) [Reference Hunter and Gaston30]. A dendrogram was constructed by entering the allele number as a character value into BioNumerics v. 6.5 (Applied Maths, Belgium) and selecting categorical coefficient and ward algorithm [Reference Lindstedt18]. By using Manhattan distance coefficient, a minimum spanning tree (MST) was generated with BioNumerics. In the MST each circle represents a different MLVA profile and the size of the circle is proportional to the number of strains in that genotype. The odds ratio (OR) with 95% confidence interval (CI) for comparing the clustering of human isolates with the clustering of isolates from mutton and animal stool samples was calculated. The dendrogram was divided into clades (strains >1 locus similarity) and MLVA-type complexes (MTCs) which were assigned for related isolates having the same allele at more than three common loci.
RESULTS
Characterization of STEC isolates
The details of dairies, total number of samples collected and percentage of isolation of STEC from different areas is shown in Table 1. A total of 250 animal stool samples and 159 meat samples were collected from hilly areas while 400 animal stool samples and 291 meat samples were collected from plane areas. We obtained 75 (12·3%) isolates from cattle stool samples, five (5%) from sheep/goat stool samples, 39 (8·6%) from mutton samples and 11 (1·8%) from human stool samples. None of the samples from chutney and chicken/pork was positive. On serotyping, 41 strains were O antigen untypable and 17 were rough. O antigen-typable strains are listed in the dendrogram (Fig. 2). Although serogroup diversity was high in STEC isolates of the non-O157 group, isolates of the same serogroup were found in samples from different sources. Figure 2 shows the geographical location and serogroup profiles of the isolates. No isolate of O157 serogroup was obtained. Table 2 shows combinations of virulence genes in STEC isolates. None of the mutton isolate was positive for the eae gene. Overall, stx2 was found to be more significantly associated with animal stool isolates compared to mutton and human stool isolates (P < 0·05) (Table 2).
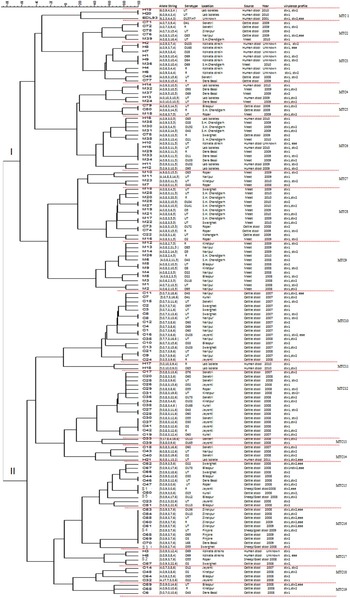
Fig. 2 [colour online]. Dendrogram based on multi-locus variable number tandem repeat analysis (MLVA) profiles of 141 non-O157 STEC strains. Dendrogram based on allelic profiles of MLVA for 141 strain including reference strain EDL933 was constructed using BioNumerics v. 6.5. Allele numbers were entered into BioNumerics as character data and the dendrogram was constructed using categorical coefficient and ward algorithm. Locus CVN002 is not been shown in the dendrogram. C, Cattle stool isolates; S, sheep/goat stool isolates; M, mutton isolates; H, human stool isolates; R, rough; UT, untypable; SH, slaughterhouse.
Table 2. Distribution of sample type, prevalence and virulence factors
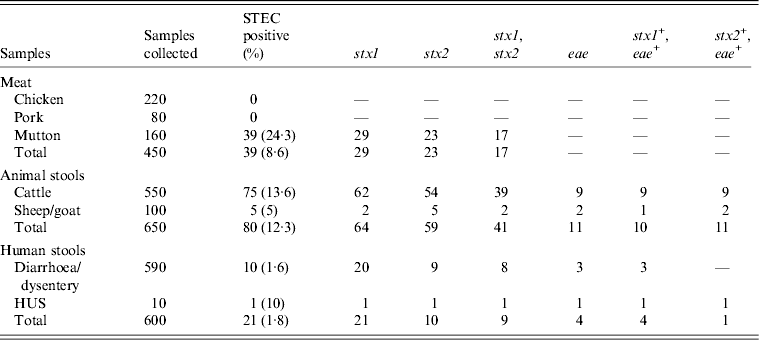
STEC, Shiga-toxin Escherichia coli; HUS, haemolytic uraemic syndrome.
MLVA genotyping
All STEC isolates were typed and 44 alleles were observed. The locus CVN002 could not be amplified in any isolate (null allele), whereas CVN003 was amplified only in six isolates. The number of alleles detected per locus ranged from three (CVN015) to 22 (CVN014). The number of repeats ranged from 1 to 16 in the six loci analysed. The DS common to all loci was 0·99.
In the dendrogram analysis, 141 isolates could be grouped into 19 different MTCs of related strains: clades I and II having nine and 10 MTCs each. Overall, 104 MLVA genotypes were observed (Fig. 2); 81 represented by one strain each and 23 shared by two or more isolates. Stool and mutton isolates formed separate lineages (clade I having mutton isolates and clade II having animal stool isolates predominantly, although few strains were interspersed). The most common genotype i.e. 5,0,9,3,7,6 was present in nine strains (eight cattle, one sheep/goat stool isolates), followed by 4,0,9,3,11,15 in five strains (two human, three mutton isolates).
Human isolates exhibited scattered distribution; 16 were present in clade I (MTCs 1, 3, 4, 6, 7) and five in clade II (MTCs 11, 14, 17). Human isolates showed a statistically significant greater OR for clustering with mutton samples (OR 8·2, 95% CI 2·6–25) compared to clustering with animal isolates (OR 0·12, 95% CI 0·04–0·37), signifying closer genetic relationship of human isolates with mutton. Serogroup-specific clustering was observed for strains M4 and M5 (serotype O22) in MTC 9; H3 and H5 (serogroup O69) in MTC 17; H1 and M36 (serogroup O69) in MTC 3. The two human isolates (H19, H20) shared a similar number of repeats with reference strain EDL933 only at two loci (CVN001, CVN007) in MTC 1 (Fig. 2). Cattle stool isolates were grouped with mutton isolates in MTCs 2, 5 and 8 while two MTCs were shared by all three types of isolates (MTCs 3 and 6). No relationship was found between the presence of stx1 and/or stx2, and the MLVA profile of the isolates.
There were only five sheep/goat isolates, that clustered with cattle stool or human isolates in MTCs 15, 16 and 17. All the five isolates shared 3–5 loci with cattle isolates and one also shared three loci with human isolates.
On the basis of allelic profiles, a MST was constructed to study the phylogeny of MLVA profiles (Fig. 3). Using the MST, the reference strain was found to be more closely related to H19 and H20 (human isolates from north India) and widely divergent from animal isolates compared to mutton isolates, further supporting the dendrogram analysis. Most of the human isolates grouped with mutton isolates except three strains (H17, H18 and H21) which clustered with animal stool isolates (Fig. 3). A few of the cattle stool isolates also grouped with mutton isolates, suggesting the contamination of mutton samples with the intestinal content of animals.
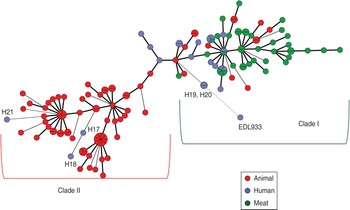
Fig. 3 [colour online]. Minimum spanning tree based on the multi-locus variable number tandem repeat analysis profiles of STEC isolates was constructed using BioNumerics v. 6.5. Animal stool isolates are depicted by red circles, mutton isolates by green circles and human stool isolates by blue circles.
DISCUSSION
Although the role of non-O157 in human disease is relatively underestimated, illnesses caused by these organisms appear to be on the rise worldwide [Reference Mathusa31]. Molecular epidemiology of STEC has not been well studied in India. MLVA was used for typing of non-O157 STEC isolated from diverse sources in a major animal rearing area of north India, with many large and small dairies, particularly in rural areas lacking proper hygiene and sanitary measures. We wanted to investigate the genetic relatedness of STEC isolated from diverse sources and whether transmission of STEC from the environment to humans was possible. We found a low prevalence of enterohaemorrhagic E. coli (EHEC) (stx1/stx2 + and eae +), similar to a previous study from India [Reference Khan25]. The prevalence rates of stx1, stx2 and eae genes (78%, 70% and 10%, respectively) were also similar to those reported from Kolkata (46%, 16% and 13·3%, respectively) [Reference Khan24]. The low prevalence of eae indicates either the absence of locus for enterocyte attaching and effacing lesions (LEE) or presence of eae variants/other adhesins in these isolates. Although many studies have shown that LEE is essential for host colonization and virulence [Reference McKee32, Reference Marches33], others have demonstrated that some STEC isolates without LEE were associated with sporadic and outbreak cases of HUS [Reference Bonnet34–Reference Newton36].
To the best of our knowledge, this is the first time STEC serogroups O2, O3, O5, O11, O21, O22, O25, O28, O30, O33, O41, O43, O52, O60, O64, O69, O85, O95, O97, O102, O110, O141, O152, O168, O172 and O173 have been typed by MLVA. New serogroups found in this study are O33, O55, O136, O165, and O173 which have not been reported previously from India. The commonest O serogroups were O43, O60, O69, and O85, which were present in seven isolates each. Serogroup O60 was isolated from animal stool only; O43 was shared between animal stool and mutton isolates whereas O85 and O69 were present in all three types of samples. Human isolates from the region belonged to serogroups O28, O64, O69, O85, O95, O102, O103, and O168. Serogroups O111, O103, O121, O145, O26, O104 and O113 are the non-O157 STEC of greatest clinical importance in many countries [Reference Noris and Remuzzi37, Reference Bosilevac and Koohmaraie38]. The EHEC isolates belonged to serogroups O85, O43, O103, O22, O173 and O136 from animal stool samples and O168, O69 from human stool samples.
In the dendrogram analysis, animal stool isolates formed a separate lineage, but a few human isolates also clustered with these, suggesting that some of the human infections may be from cattle, perhaps through milk, animal contact or the environment. As humans and animals live in close proximity, direct faecal–oral transmission is also a possibility in our area. Outbreaks of O157 have been reported from visits to farms, petting zoos and fairs [Reference Tozzi, Gorietti, Caprioli, Duffy, Garvey and McDowell39]. Even though we found different serogroups sharing the same MLVA profiles, there is no clear evidence of transmission from bovine to humans as one serogroup may have many serotypes and also, strains had different virulence profiles. Sixteen of the human isolates clustered with mutton samples, thus suggesting genetic relatedness between mutton and human isolates. The consumption of contaminated mutton is an important source of transmission to humans [Reference Elder40]. An interesting finding was that only a small number of cattle strains clustered with mutton or human strains, demonstrating that only a few of these strains have the potential for further transmission. The presence of large volumes of animal manure in the environment acts as a potential reservoir of enteric pathogens [Reference Kudva41]. Out of 15 EHEC isolates, 11 were isolated from animal stool, signifying the presence of an environmental reservoir of isolates that are potentially pathogenic to humans and hence, may have public health importance. The cattle and sheep/goat isolates clustered together due to the rearing of these animals on the same premises. None of the sheep/goat stool isolates clustered with mutton isolates signifying that mutton samples were becoming contaminated with cattle stool isolates due to unhygienic practices, and some of these cattle stool isolates belong to a lineage that is able to be further transmitted.
One of the major limitations of our study was non-availability of isolates from outbreaks of STEC illness. Such outbreaks have not been reported from India. Further, the environment sampling was not epidemiologically linked to human cases. Another limitation was the inability to perform H typing. Although some previous studies from India have reported the isolation of O157 from food, water and cattle [Reference Manna42–Reference Surendraraj44], this serotype was conspicuously absent from all sources in this study, indicating the low prevalence of this organism in this geographical region. In the North Indian region beef consumption is low due to religious beliefs. Thus, we were not able to collect beef samples. However, this does not explain the absence of this serotype in animal stool samples. Immunomagnetic separation technique, using anti-E. coli O157 (Invitrogen, Dynal AS, Norway) may be more sensitive to detect O157.
CONCLUSION
This is the first study from India in which MLVA was used to genotype non-O157 STEC from diverse sources. Although animal stool and mutton isolates formed separate lineages, the majority of human isolates clustered with those from mutton, and only a few human isolates clustered with animal stool isolates. Further epidemiological studies are required to investigate these sources in relation to the occurrence of human cases.
ACKNOWLEDGEMENTS
We gratefully acknowledge Dr Mani Kavaratty Raju, Director, National Salmonella and Escherichia Centre, Central Research Institute, Kasauli 173204 (India), for serogrouping of the isolates. We also acknowledge Dr G. B. Nair from the National Institute of Cholera and Enteric Diseases for providing isolates of STEC from Kolkata. We thank Dr Shivaprakash for designing the dendrogram and MST using BioNumerics v. 6.5. We gratefully acknowledge the kind help provided by Dr Ramesh Chand for grammatical correction of the manuscript.
The authors acknowledge the financial, administrative and technical support from the Indian Council of Medical Research (ICMR), New Delhi, India (grant no. 5/8-1(213)-D/2006 ECD-II).
DECLARATION OF INTEREST
None.








