Introduction
The parasite Leishmania major is the causative agent of cutaneous leishmaniasis in the Old World. Leishmania survives and replicates within host macrophages and evades numerous host defences by inhibition of important cell functions, including the modulation of host cell signalling pathways through phosphorylation and dephosphorylation mechanisms. Protein phosphorylation and dephosphorylation processes are regulated by protein kinases and phosphatases, respectively, which generally occur on tyrosine, serine or threonine residues (Cohen, Reference Cohen2002; Wang et al. Reference Wang, Zhang and Wei2008). Protein phosphatases are classified in three families based on substrate specificity, sequence alignment, structure and their catalytic mechanism, which are: PPP (phosphoprotein phosphatases), PPM (metal-dependent protein phosphatases) and PTP (protein tyrosine phosphatases) (Moorhead et al. Reference Moorhead, Trinkle-Mulcahy and Ulke-Lemee2007). The PPP family consists of the phosphatases PP1, PP2A and PP2B. The PPM family contains type 2C protein phosphatases (PP2C) and pyruvate dehydrogenase phosphatase (PDP). The PP2C is a monomeric enzyme that requires the metal cations Mg+2 or Mn+2 for enzymatic activity (Das et al. Reference Das, Helps, Cohen and Barford1996; Andreeva and Kutuzov, Reference Andreeva and Kutuzov2004; Schweighofer et al. Reference Schweighofer, Hirt and Meskiene2004; Lammers and Lavi, Reference Lammers and Lavi2007; Moorhead et al. Reference Moorhead, Trinkle-Mulcahy and Ulke-Lemee2007). In the human genome, 16 genes have been identified which generate 22 isoforms by a process of alternative splicing; each of these enzymes regulates distinct signalling pathways (Das et al. Reference Das, Helps, Cohen and Barford1996). The roles of PP2C in eukaryotic cells are diverse and have been described in different organisms such as the plant Arabidopsis thaliana, in which 76 candidates of PP2C-type phosphatases were identified in its genome (Schweighofer et al. Reference Schweighofer, Hirt and Meskiene2004). Different PP2C from A. thaliana participate in various signalling pathways, such as the one regulated by abscisic acid and the signalling pathways activated by mitogen protein (Mitula et al. Reference Mitula, Tajdel, Ciesla, Kasprowicz-Maluski, Kulik, Babula-Skowronska, Michalak, Dobrowolska, Sadowski and Ludwikow2015). In protozoan parasites such as Plasmodium falciparum, an unusual PP2C phosphatase (PfPP2C) was identified. It has two catalytic sites, one with serine and the other with threonine, with an identity between them of about 9% (Mamoun et al. Reference Mamoun, Sullivan, Banerjee and Goldberg1998). Sporozoites of Plasmodium yoelli and Plasmodium berguei, incubated with increasing concentrations of potassium, enhanced the infectivity of the parasites and the copy number of the transcripts for PP2C was increased, when compared with parasites not exposed to potassium in which the expression of the enzyme was kept at basal levels (Kumar et al. Reference Kumar, Garcia, Chandran, Van Rooijen, Zhou, Winzeler and Nussenzweig2007). Toxoplasma gondii also has a protein phosphatase PP2C (TgPP2C), which is secreted from the rhoptries, a group of secretory organelles involved in host cell invasion. Once inside the parasitophorous vacuole, TgPP2C is transported to the nucleus of the host cell. In contrast, a PP2C knockout strain had reduced intracellular growth, which was reversed by complementation with the wild-type gene for PP2C (Gilbert et al. Reference Gilbert, Ravindran, Turetzky, Boothroyd and Bradley2007). PP2C was cloned in the parasite Leishmania chagasi (LcPP2C); however, there are no reports of its function in the parasite or in the host cell (Burns et al. Reference Burns, Parsons, Rosman and Reed1993). The genome analysis of L. major predicted the existence of 45 phosphatases of the PPP family and 15 of the PPM family; however, the biochemical characterization and localization of these enzymes in this parasite have not yet been reported. We previously identified a membrane-bound PTP in L. major that differs in expression, activity and ultrastructural localization between the procyclic and metacyclic stages of the parasite life cycle (Aguirre-Garcia et al. Reference Aguirre-Garcia, Escalona-Montano, Bakalara, Perez-Torres, Gutierrez-Kobeh and Becker2006). We now report the cloning, expression, purification and biochemical characterization of PP2C from L. major. The enzyme was identified in different Leishmania species and its ultrastructural localization is shown in L. major promastigotes.
Materials and methods
Culture of Leishmania spp promastigotes
Leishmania major MHOM/SU/73/5-ASKH (a generous gift from Dr H. Moll, University of Würzburg, Germany), L eishmania (L.) mexicana MNYC/BZ/62/M379, L eishmania (L.) venezuelensis MHOM/VE/80/PMH3, L eishmania (L.) amazonensis IFLA/BR/67/PH8, L eishmania (V.) panamensis MHOM/PA/71/LS94, L eishmania (V.) brazilienzis MHOM/BR/75/M2903, L eishmania (V.) donovani, [a generous gift from Dr Nancy Sarabia, Centro Internacional de Entrenamiento e Investigación Médica in Calí, Colombia (CIDEIM)], promastigotes were cultivated in RPMI 1640 medium, which contained penicillin (100 units mL−1), streptomycin (100 µg mL−1) and 10% fetal bovine serum, at 26 °C (all reagents were from Gibco, Invitrogen Corporation Carlsbad, CA, USA). The cultured promastigotes were washed twice with phosphate-buffered saline (PBS) at pH 7·5, centrifuged for 10 min at 2500 × g at room temperature (RT).
Extraction of DNA and amplification of the gene for PP2C from L. major
Total DNA was extracted from L. major promastigotes using TRIZOL. The gene was amplified using the forward oligonucleotide (Fw) 5′ATGGGCATTCCACTTCCGA3′ and reverse oligonucleotide (Rv) 5′TCACTGCGTCTGCTCACC3′. The PCR was performed using 100 µL of the following reaction mixture: 1.5 mm MgCl2; 0.2 mm deoxynucleoside triphosphates; 100 ng of the corresponding oligonucleotides; 10 ng genomic DNA and one unit of Taq DNA polymerase (Invitrogen) using 40 cycles for 5 min at 94 °C, 30 s, 1 min at 50.7 °C, 30 s and 48 s at 72 °C, and a final extension of 10 min at 72 °C. The gene was cloned into pET-23b expression plasmid using the Nde l and Hind III restriction sites. The gene was completely sequenced and transformed into BL21 (DE3) pLysS cells (Novagen, Madison, WI, USA). Bacteria containing the plasmid with the gene of protein phosphatase PP2C of L. major were grown in Luria-Bertani medium and selected for ampicillin resistance (100 µg mL−1) at 37 °C allowing them to grow until the OD of the culture at 600 nm reached 0·8–1·0. Expression of the recombinant protein was induced with 0.4 mm isopropyl-thio-b-D-galactopyranoside (IPTG) for 3 h. After incubation, bacteria were harvested by centrifugation and processed immediately, or frozen at −70 °C until use.
Purification of recombinant PP2C from L. major (LmPP2C)
Cell cultures in volumes of 100 mL were centrifuged and suspended in 50 mL lysis buffer (50 mm Tris HCl, pH 8; 300 mm NaCl; 1 mm benzamidine, 100 µm leupeptin, 2 µg mL−1 aprotinin and 5 mm imidazole). The suspension was sonicated five times for 40 s with resting periods of 2 min between each interval and 32% amplitude, using a ModelVCX 650 Ultrasonic processor (Ultrasonics, Inc.). The homogenate was centrifuged at 21 000 × g for 1 h at 4 °C to obtain the supernatant with the soluble protein. The supernatant was loaded onto a Ni-charged column previously equilibrated with binding buffer (50 mm Tris-HCl, pH 8; 300 mm NaCl and 5 mm imidazole). The recombinant protein (LmPP2C) was purified and eluted with an elution buffer (50 mm Tris-HCl, pH 8; 300 mm NaCl and 50–500 mm imidazole) and the protein concentration was quantified using the Bradford method (Bradford, Reference Bradford1976).
Production of polyclonal antiserum
Anti-LmPP2C antibodies were generated in rabbits as previously described by Montfort and collaborators (Montfort et al. Reference Montfort, Perez-Tamayo, Perez-Montfort, Gonzalez Canto and Olivos1994). Before the immunization, the rabbits were bled, and pre-immune serum was collected. Briefly, rabbits were injected intramuscularly with 150 µg of recombinant LmPP2C emulsified in complete Freund's adjuvant; the same procedure was repeated 2 weeks later without adjuvant. The immunization was done with two weekly intramuscular injections, after which the animals were bled and antiserum was separated by centrifugation and stored at −20 °C. The pre-immune and immune sera were used for Western blot analysis. Rabbits were housed at the animal facilities of the Unidad de Investigación en Medicina Experimental de la Facultad de Medicina, UNAM, and their handling was done following the National Ethical Guidelines for Animal Health NOM-062-ZOO-1999 and the guidelines recommended for animal care by the Ethical Committee of the Medical School of the UNAM.
Phosphatase activity assays
p-NPP substrate
Acid phosphatase activity was determined as described by Dissing et al. (Reference Dissing, Dahl and Svensmark1979). Briefly, 0·7 µg of recombinant LmPP2C was incubated in the following buffers: 50 mm MES, pH 5–6·5; 50 mm MOPS, pH 7–7·5; 50 mm HEPES, pH 8; 50 mm TRIS, pH 8·5–9 and 50 mm CAPS, pH 10–11 and added to 10 mm MgCl2 and 10 mm p-nitrophenyl phosphate [p-NPP] in a final volume of 100 µL for 60 min at 37 °C. Afterwards, the reaction was stopped with 20 µL of 2 N NaOH. The absorbance was read at 405 nm using a micro titre plate reader. Divalent cations (10 mm MgCl2 or MnCl2) were added in 50 mm HEPES, pH 8.
Phosphopeptides
Tyrosine and serine/threonine phosphatase activity was assayed using Promega's non-radioactive tyrosine phosphatase assay system. The release of inorganic phosphate (P i ) was monitored by measuring the absorbance of the molybdate-malachite green-phosphate complex. A 0·7 µg of recombinant LmPP2C was incubated in a total volume of 100 µL of assay buffer containing 50 mm HEPES pH 8·0 plus 10 mm MgCl2. The reaction was started by adding 50 µm Tyr phosphopeptide-1 substrate [END (pY) INASL], 50 µm Thr [RRA (pT)VA] during 30 min at RT and stopped with 50 µL molybdate dye/additive mixture. The optical density of the samples was read at 630 nm, using a curve of phosphates as standard (Aguirre-Garcia et al. Reference Aguirre-Garcia, Escalona-Montano, Bakalara, Perez-Torres, Gutierrez-Kobeh and Becker2006).
Effect of inhibitors of phosphatases on recombinant LmPP2C
The activity of recombinant LmPP2C (0·7 µg) was analysed in the presence of specific PTP inhibitors such as 200 µm sodium orthovanadate, 200 µm ammonium molybdate and 50 µm sodium pervanadate. Additionally, serine/threonine phosphatase inhibitors were tested such as 5 nm calyculin and 1 µm okadaic acid.
Sanguinarine, a specific inhibitor for PP2C, was used at 0·1, 0·5, 1, 5, 10 and 20 µm. For the inhibition assays, 100 µL of the reaction mixture was pre-incubated for 15 min at RT before adding the p-NPP substrate (all reagents from Sigma-Aldrich, St. Louis, MO, USA) and were incubated thereafter for another 60 min at 37 °C. Afterwards, the reaction was stopped with 20 µL of 2 N NaOH. The absorbance at 405 nm was read using a micro titre plate reader. Recombinant PTP from Yersinia enterocolitica (YePTP) was used as a control of PTP activity (purchased from Calbiochem, La Jolla, CA, USA).
SDS-PAGE and Western blot
Identification of the His-Tag in recombinant LmPP2C
Total extract (TE) of bacteria was analysed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) in 10% acrylamide gels and then electro transferred to nitrocellulose membranes. The blots were incubated with a polyclonal HRP anti-His6-Tag antibody at a 1:1000 dilution in TBS-T (200 mm Tris-HCl, 150 mm NaCl, 0·005% Tween-20) and washed four times every 10 min with TBS-T.
Identification of PP2C in TEs of L. (L.) mexicana, L. (V.) panamensis, L. (V.) brazilienzis, L. (L.) venezuelensis, L. (V.) donovani, L. (L.) amazonensis, L. major promastigotes
All promastigotes were harvested by centrifugation at 2000 × g for 10 min and then washed three times with PBS. The pellet containing the parasites was suspended in cold lysis buffer (10 mm imidazole pH 7·2, 2 µg mL−1 leupeptin, 10 µg mL−1 aprotinin, 2 mm benzamidine) and then sonicated (TE). Afterwards, 10 µg of TE from different Leishmania species and were analysed by SDS-PAGE in 10% acrylamide gel and electro transferred onto nitrocellulose membranes. The PP2C protein was used as a control. These were washed with TBS-T and blocked by treatment with 5% milk in TBS-T for 1 h; then they were incubated with antibodies against LmPP2C using a dilution of 1:1000 in 1% milk in TBS-T overnight at 4 °C. After this incubation, the membranes were washed with TBS-T and incubated with the secondary HRP-goat anti-rabbit IgG antibody (Biomeda, Foster City, CA, USA) at a dilution of 1:5000 in TBS-T with 3% of milk. The membranes were finally washed 10 times in TBS-T.
Bands were detected using enhanced chemiluminescent substrate (Super-Signal West Pico Chemiluminescent Substrate, Pierce, Rockford, IL, USA), following the manufacturer's instructions.
Fluorescence microscopy
Leishmania major promastigotes were washed with PBS and allowed to adhere to poly-L-lysine-coated cover slides for 90 min. They were then fixed with 2% paraformaldehyde in PBS for 30 min at RT and washed three times with PBS. Afterwards, these cells were permeabilized with 5% triton X-100 and blocked with 5% BSA for 30 min. They were then incubated overnight at 4 °C with antibodies against the recombinant LmPP2C using a 1:100 dilution. The parasites were washed with PBS after the incubation and stained with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG secondary antibodies at a 1:200 dilution. Cells were counterstained with 4′, 6′-2 diamidino-2-phenylindole (DAPI) at a 1:1000 dilution in PBS. Cover slides were mounted with vectashield mounting medium (Vector Laboratories, Inc., Burlingame, CA, USA) and observed in a fluorescence microscope (Axioskop 2 mot plus, Carl Zeiss, Germany). The images were recorded in an Axiocam HR digital camera and processed with the Axiovision software (Carl Zeiss, Version 4.8), (Gomez de Leon et al. Reference Gomez de Leon, Diaz Martin, Mendoza Hernandez, Gonzalez Pozos, Ambrosio and Mondragon Flores2014).
Immunolocalization of PP2C by immune electron microscopy (IEM)
Detection of PP2C in L. major promastigotes was achieved by electron microscopy (Gomez de Leon et al. Reference Gomez de Leon, Diaz Martin, Mendoza Hernandez, Gonzalez Pozos, Ambrosio and Mondragon Flores2014). Briefly, promastigotes were washed with PBS and then fixed with a mixture of 4% paraformaldehyde and 0·1% glutaraldehyde in PBS for 1 h at RT. Once washed, the promastigotes were gradually dehydrated in ethanol and embedded in LR White resin (London Resin, Polysciences, Inc., Warrington, PA, USA). Promastigotes in resin were deposited in plastic moulds and were polymerized overnight, under ultraviolet light, at 4 °C. Thin sections of the blocks were obtained in an Ultracut E ultra-microtome (Reichert Jung, Austria) and then mounted on Formvar-covered nickel grids. Immunogold labelling was carried out by flotation of the mounted sections on drops of the respective solutions; non-specific labelling was diminished by incubation with 1% skim milk and PBS-T (PBS and 0·05% Tween-20) for 30 min. Grids with the sections were incubated with the antibody against LmPP2C at a 1:10 dilution in PBS-T for 1 h at RT and overnight at 4 °C. Grids were washed with PBS-T and then incubated with a goat anti-rabbit polyclonal antibody coupled to 10 nm gold particles for 2 h at RT (Zymed, Thermo Scientific, PA, USA) (1:40 dilution in PBS-T). After thorough washings in PBS and distilled water, sections were contrasted with 2% uranyl acetate and a saturated solution of lead citrate and then examined with a transmission electron microscope (TEM, JEOL 1400x, JEOL Ltd, Japan). As negative control, sections were incubated with pre-immune rabbit serum diluted in PBS-T and then with the secondary antibody coupled to gold particles.
The identification of the subcellular structures positive for PP2C was performed using thin sections of parasites that were processed to preserve the ultrastructure. (Gomez de Leon et al. Reference Gomez de Leon, Diaz Martin, Mendoza Hernandez, Gonzalez Pozos, Ambrosio and Mondragon Flores2014). Briefly, promastigotes were fixed for 1 h in 2·5% glutaraldehyde. After thorough rinsing in PBS, parasites were post-fixed for 1 h in 1% OsO4 at 4 °C, rinsed, gradually dehydrated in ethanol, and finally embedded in Spurr's resin. Thin sections were obtained with an Ultracut E ultramicrotome and stained with uranyl acetate and lead citrate. Copper grids with the sections were examined in the TEM at 80 keV. Digital images were obtained and processed with Adobe Photoshop software (USA).
Bioinformatic analysis
Homology modelling was performed using the SWISS-MODEL server (http://swissmodel.expasy.org/interactive); the model was validated with the Mod Eval software version master.e8f19a7 (https://modbase.compbio.ucsf.edu/evaluation//), and the structure obtained was edited using the Swiss-PDBViewer programme version 4.1 (http://www.expasy.org/spdbv/). Secondary structure prediction was carried out using PSIPRED Protein Sequence Analysis Workbench version 3.3 (http://bioinf.cs.ucl.ac.uk/psipred/) (Melo et al. Reference Melo, Sanchez and Sali2002; Arnold et al. Reference Arnold, Bordoli, Kopp and Schwede2006; Shen and Sali, Reference Shen and Sali2006; Bordoli et al. Reference Bordoli, Kiefer, Arnold, Benkert, Battey and Schwede2009; Guex et al. Reference Guex, Peitsch and Schwede2009; Buchan et al. Reference Buchan, Minneci, Nugent, Bryson and Jones2013; Biasini et al. Reference Biasini, Bienert, Waterhouse, Arnold, Studer, Schmidt, Kiefer, Gallo Cassarino, Bertoni, Bordoli and Schwede2014).
Phylogenetic analysis of L. major PP2Cs
The non-rooted phylogenetic tree was constructed using the MEGA 6 programme from the alignment of 15 amino acid sequences PP2Cs present in the L. major genome with the protein sequence of the LmxM.25.0.750 gene of L. mexicana and the protein sequence of the human PP2C (PPM1A, NP_066283.1), which was used as seed.
Results
Cloning, expression and purification of PP2C of L. major
In order to determine the primary structure of PP2C homologue from L. major, forward and reverse primers were constructed from the sequence of the LmjF.25.0750 gene in TriTrypDB, the database of Trypanosomatids (tritrypdb.org). A fragment of 1200 bp was amplified from L. major DNA using PCR. The pET-23b (+) expression vector containing a C-terminal His6 Tag was transformed into BL21 strain of Escherichia coli for expression and purification of the PP2C fusion protein. The transformed BL21 cells were grown and induced with 1 mm IPTG for 3 h at 30 °C, the recombinant protein was observed in gels stained with Coomassie blue (Fig. 1A, lanes 2 and 3), in contrast to cells in which the expression of the protein was not induced (Fig. 1A, lane 1). The presence of the histidine tag in the recombinant protein was verified by Western blot with specific antibodies that recognized the C-terminal His6 Tag (Fig. 1B, lanes 2 and 3), which was negative in non-induced cells (Fig. 1B, lane 1). TE of L. major promastigotes and recombinant protein were analysed by SDS-PAGE and stained with Coomassie blue (Fig. 1C, lanes 1 and 2). This result showed that the purified protein had the expected molecular mass of 44·9 kDa. The anti-LmPP2C antibodies were checked by Western blot using TE of L. major and purified LmPP2C. These antibodies recognized a 44·9 kDa protein in both samples (Fig. 1D, lanes 1 and 2).
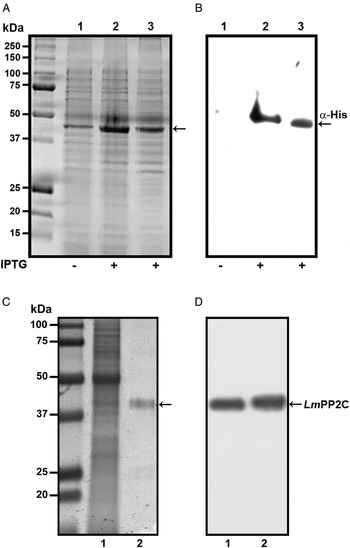
Fig. 1. Expression, purification and identification of PP2C of L eishmania major (Lm PP2C). (A) Expression of Lm PP2C in different fractions of Escherichia coli cells. Crude extract prepared from non-induced E. coli cells BL21 (1), E. coli cells induced with IPTG (2) and supernatant (3). SDS-PAGE gel was stained with Coomassie-blue dye. (B) Western blot analysis of the same sample used above: crude extract prepared from non-induced E. coli cells BL21 (1), E. coli cells induced with IPTG (2) and supernatant (3). The C-terminal His-Tag was detected using a monoclonal antibody. (C) SDS-PAGE gel of TE of L. major (1) and Lm PP2C purified (2) were stained with Coomassie Blue. (D) Western blot of TE of L. major (1), and purified Lm PP2C (2), using polyclonal antibodies against Lm PP2C.
Multiple sequence alignment
Using Clustal W multiple-sequence alignment, the sequence of the cloned LmPP2C revealed that it has 98% similarity with the sequence of PP2C of L. chagasi. It also demonstrated that the complete LmPP2C open reading frame shares 11 distinct conserved motifs in the enzymes belonging to this family (Fig. 2A).
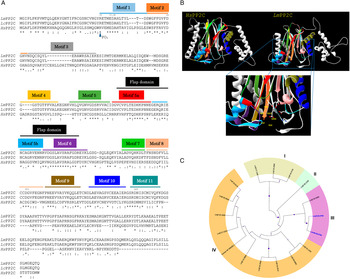
Fig. 2. Multiple sequence alignments of human, L eishmania major and L eishmania chagasi PP2C and phylogenetic analysis of L. major PP2Cs. (A) Multiple sequence alignment of Human PP2C (HsPP2C), L. major (LmPP2C) and L. chagasi (LcPP2C). Alignment of the catalytic domain sequences was performed using the programme CLUSTAL W. The most important residues of the three PP2C are indicated with numbers and motifs with colours: motif 1 (blue line), motif 2 (orange line), motif 3 (grey line), motif 4 (yellow line), motif 5 (dark green line), the Flap domain is indicated by a black line. Motifs 5a and 5b are shown with a red and dark blue line respectively, motif 6 (purple line), motif 7 (clear green line), motif 8 (pink line), motif 9 (brown line), motif 10 (blue line) and motif 11 (water green line). (B) Ribbon diagram of HsPP2C and LmPP2C. The catalytic site of LmPP2C was amplified, showing the important amino acids. (C) Phylogenetic analysis of L. major PP2Cs. The non-rooted phylogenetic tree was constructed in the MEGA 6 programme from the alignment of 15 amino acid sequences PP2Cs present in the L. major genome with the protein sequence of the LmxM.25.0.750 gene of Leishmania mexicana and the protein sequence of the human PPM1A (NP_066283.1). Seventeen proteins were grouped into four groups and designated with different colour (I–IV). The evolutionary history was inferred using the maximum probability, method based on the Le_Gascuel_2008 model.
Furthermore, both HsPP2C and LmPP2C have conserved amino acids for catalytic activity such as: the aspartic acid residues involved in the coordination of Mg+2 and Mn+2 cations. The catalytic site of PP2C is localized in a cleft between two central β sheets formed by aspartic acids D38, D55, D221, D265, arginine R33 and glutamic acid E37 (Fig. 2B). Identical residues were identified at these positions in all aligned sequences (Fig. 2A). These residues were confirmed to be present in LmPP2C.
Phylogenetic tree analysis
To compare the protein LmjF.25.0750 analysed in this work with the members of the PP2C in L. major family and for the identification of conserved residues, a multiple-sequence alignment was constructed using the 15 L. major PP2C, LmxM.25.0750 and the human PP2C (PPM1A). Similarity relationships among the amino acids sequences were obtained and we identified characteristic motifs preserved by evolution. A total of 11 conserved signature motifs were identified that seem to correspond to structural elements and exposed functional residues of PP2C proteins (data not shown). To investigate the phylogenetic relationships of PP2C genes between L. major phosphatases, L. mexicana PP2C and the human PPM1A, we constructed a phylogenetic tree based on the alignments of PP2C domains using the maximum likelihood method. The phylogenetic analyses indicated that the 15 LmPP2C proteins were divided into four groups (Fig. 2C, I–IV). The LmjF 25.0750 recombinant protein clustered together with LmxM.25.0750 (Fig. 2C, III), while the nine Lmj phosphatases formed an independent group (Fig. 2C, IV). There was a cluster of three proteins in group I, whereas LmjF 36.1230 was in an independent branch (Fig. 2C, II).
Biochemical characterization of LmPP2C
Cation dependence
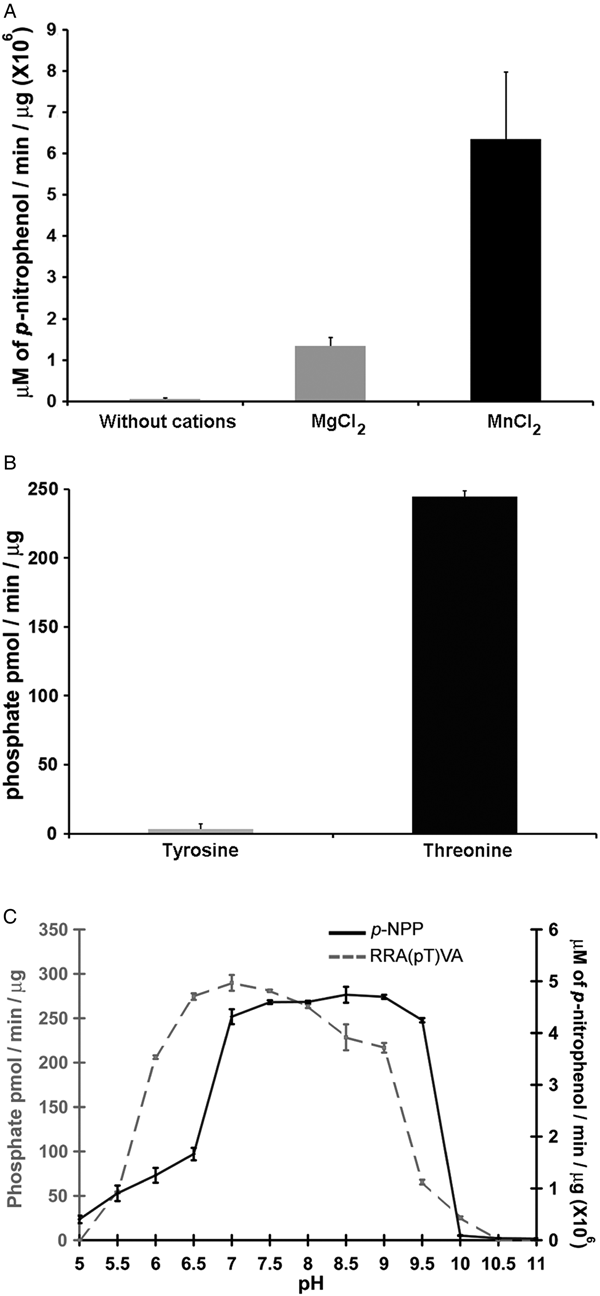
Phosphatases PP2C are usually monomeric enzymes that require metal cations such as Mg+2 or Mn+2 for their enzymatic activity. We analysed the activity of LmPP2C with the substrate p-NPP in the presence or absence of divalent cations. We observed that LmPP2C showed higher phosphatase activity in the presence of MnCl2, compared with MgCl2 (Fig. 3A).
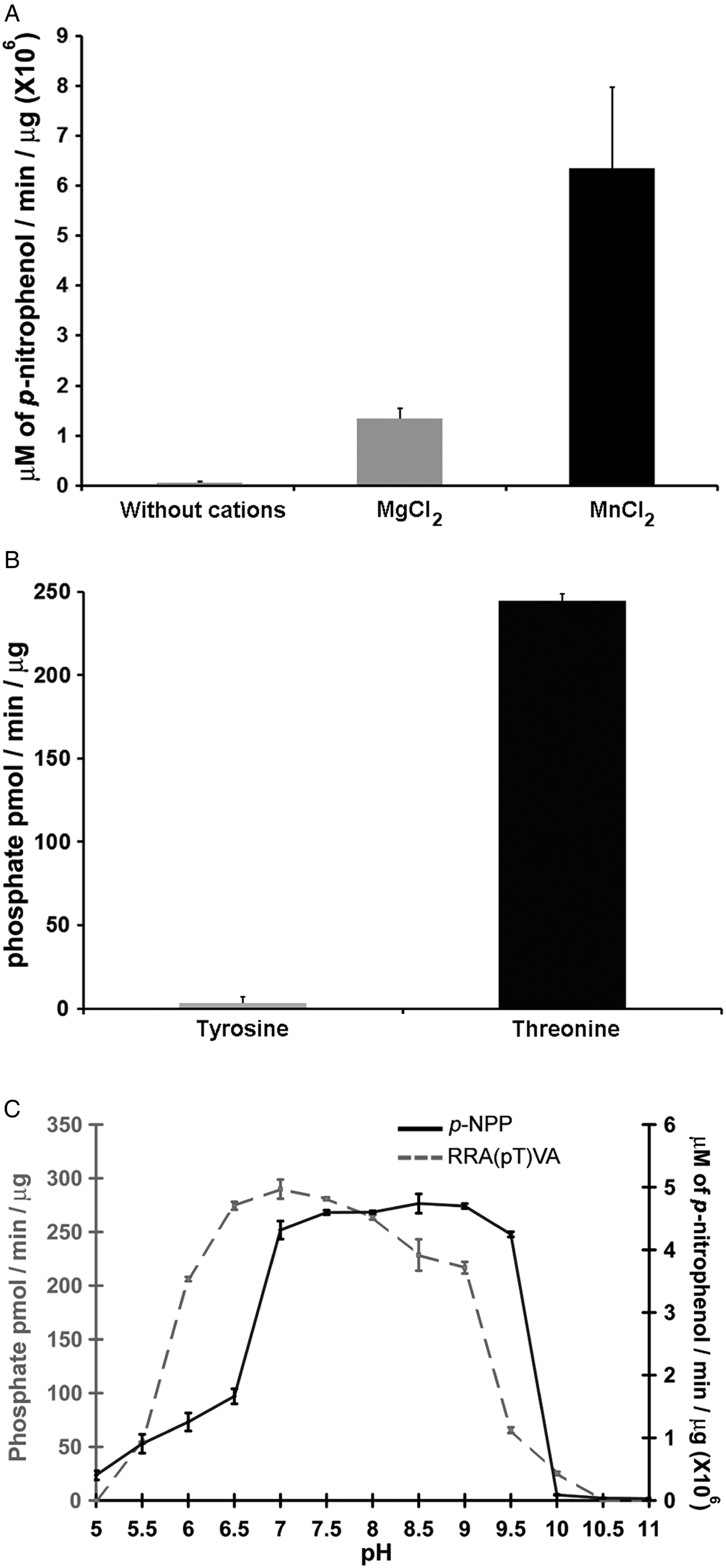
Fig. 3. Biochemical characterization of LmPP2C. (A) The effect of MgCl2 and MnCl2 on LmPP2C activity was measured under standard conditions using p-NPP 10 mm as a substrate. (B) Hydrolysis of [END (pY) INASL] and Thr [RRA (pT) VA] by LmPP2C. (C) Determination of optimum pH for the activity of LmPP2C. PP2C was incubated with the appropriate buffers at different pH (5–11) in the presence of 10 mm MgCl2 and different substrates: p-NPP and RRA(pT) VA.
Substrate specificity
Substrate specificity was tested by dephosphorylation assays using peptides phosphorylated in tyrosine and threonine residues. It was observed that LmPP2C dephosphorylate preferentially the threonine substrate (Fig. 3B), whereas the tyrosine substrate was not dephosphorylated.
Optimum pH
The influence of pH on the enzymatic activity for recombinant LmPP2C was evaluated using two different substrates: p-NPP and the phosphothreonine peptide [RRA (pT) VA] in different buffers. The results showed that the optimal activity of PP2C phosphatase reached at pH 8·5 using p-NPP as substrate, while the phosphothreonine substrate showed the maximum at pH 7 (Fig. 3C).
Effect of various inhibitors of phosphatases on the activities of LmPP2C
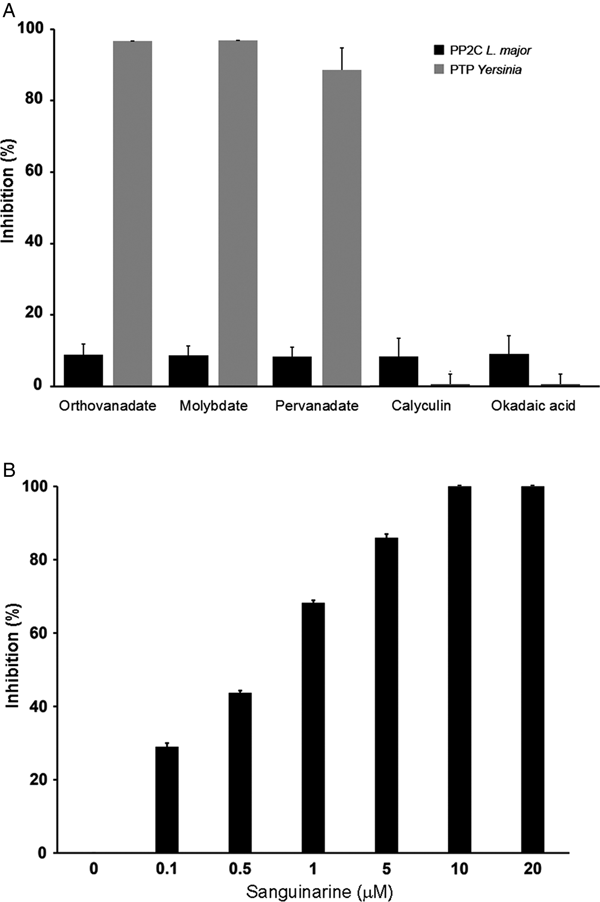
In order to characterize if the phosphatase activity is due to PTP or PP2C activity, we used different phosphatase inhibitors: orthovanadate, molybdate and pervanadate for PTP and calyculin, okadaic acid and sanguinarine for serine/threonine phosphatases.
LmPP2C was not inhibited by the specific PTP inhibitors (Fig. 4A), which contrasts with the YePTP that was completely inhibited by these compounds (Fig. 4A). Calyculin and okadaic acid did not inhibit LmPP2C or YePTP (Fig. 4A). However, sanguinarine, the specific inhibitor of PP2C activity, inhibited LmPP2C in a dose-dependent manner (0·1–20 µm), with a complete inhibition at a concentration of 10 µm (Fig. 4B).
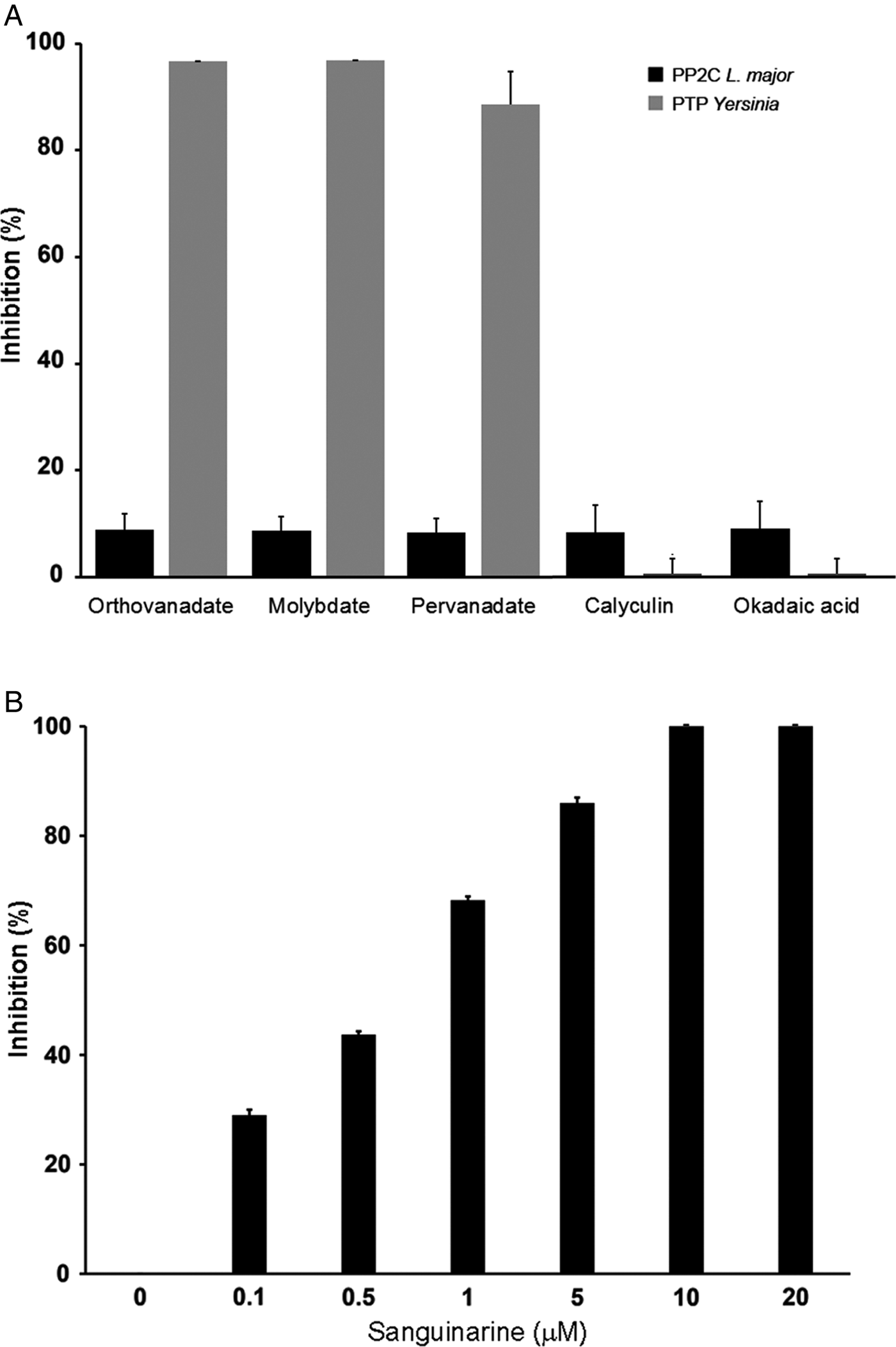
Fig. 4. Effect of different inhibitors on LmPP2C and YePTP activities. (A) Inhibitors of PTP (orthovanadate, molybdate and pervanadate) and inhibitors of serine threonine phosphatases (calyculin and okadaic acid) were incubated with LmPP2C, and YePTP and phosphatase activity was analysed by p-NPP. (B) An inhibitor of the PPM family (sanguinarine) was incubated at different concentrations with LmPP2C. The bars represent the mean values of the per cent relative activity obtained separately in three different experiments.
Western blot reactivity of recombinant LmPP2C
Antibodies were prepared against the recombinant LmPP2C (α-LmPP2C) and used to identify the PP2C protein in TE promastigotes from different Leishmania species: L. (L.) mexicana, L. (V.) panamensis, L. (V.) brazilienzis, L. (L.) venezuelensis, L. (V.) donovani, L. (L.) amazonensis, L. major and LmPP2C. In all Leishmania species, a 44·9 kDa protein was identified (Fig. 5A, lanes 1–7) which had the same molecular mass as LmPP2C (Fig. 5A, lane 8). Tubulin, as a loading control, was included in the same experiment.
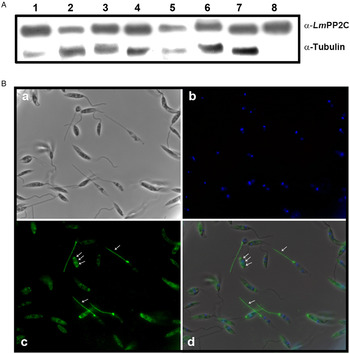
Fig. 5. PP2C: immunodetection in TE promastigotes from different Leishmania species and immunolocalization in promastigotes. (A) Immunodetection of PP2C protein in TE promastigotes from L eishmania (L.) mexicana (1), L eishmania (V.) panamensis (2), L eishmania (V.) brazilienzis (3), L eishmania (L.) venezuelensis (4), L eishmania (V.) donovani(5), L eishmania (L.) amazonensis (6), L eishmania major (7), promastigotes Lm PP2C recombinant was used as control (lane 8). Tubulin as a loading control. (B) Immunolocalization of PP2C in L. major promastigotes by immunofluorescence microscopy. Parasites were observed in phase contrast (panel a), the nucleus was stained with DAPI (panel b). Distribution of Lm PP2C was determined by using an antibody against PP2C and then evidenced with a secondary antibody conjugated to FITC (panel c). Inset d represents the merge of the different staining conditions. Scale bar = 5 µm.
Immunolocalization of PP2C in L. major promastigotes by immunofluorescence
The analysis of the distribution of the PP2C in promastigotes of L. major was performed by indirect immunofluorescence. Parasites were observed using phase contrast microscopy (Fig. 5B, panel a). The nucleus was stained with DAPI (Fig. 5B, panel b). In several parasites, the distribution of PP2C showed a fluorescent pattern confined to the flagellum and to the flagellar pocket (big arrows). Interestingly, some parasites exhibited a three-band pattern located in the cell body (small arrows) without staining of the flagella (Fig. 5B, panel c). The merge of both: nuclei and PP2C labelling, is shown in panel d (Fig. 5B). The negative control with pre-immune serum showed no labelling of the parasite (data not shown).
Ultrastructural localization of PP2C in promastigotes of L. major
The distribution of PP2C in promastigotes of L. major, studied by IEM using α-LmPP2C, showed labelling of the flagellum and the flagellar pocket (Fig. 6C–E). In order to determine precise location of the immune gold labelling, fine structure images of the flagellar pocket and the flagellum were done in longitudinal and transverse sections (Fig. 6A and B). Negative controls with pre-immune serum showed no labelling of any structure of the parasite (data not shown).
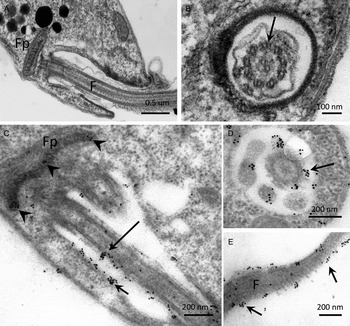
Fig. 6. IEM localization of LmPP2C in promastigotes. Insets (A) and (B) correspond to controls of ultrastructure of the flagellar pocket of promastigotes seen in longitudinal and transverse thin sections, respectively. Insets (D, E) correspond to the immuno-gold labelling of the LmPP2C in magnified zones of the promastigote in longitudinal (inset C) and transverse (inset D) views of the flagellar pocket. Inset (E) corresponds to the labelling of LmPP2C in a magnified zone of the flagellum. Scale bar = 200 nm.
Discussion
In recent years, the number of PP2C family members has grown steadily, and more interesting features, phenotypes and potential clinical applications have been described for the family of phosphatases. Orthologs of human PP2C can be found in virtually all organisms, ranging from mammals, insects, plants, bacteria and yeasts to parasites. This conservation throughout evolution indicates that these enzymes probably play important roles in regulating key cellular signalling events (Su and Forchhammer, Reference Su and Forchhammer2013). The genome of L. major was reported in 2005 (Ivens et al., Reference Ivens, Peacock, Worthey, Murphy, Aggarwal, Berriman, Sisk, Rajandream, Adlem, Aert, Anupama, Apostolou, Attipoe, Bason, Bauser, Beck, Beverley, Bianchettin, Borzym, Bothe, Bruschi, Collins, Cadag, Ciarloni, Clayton, Coulson, Cronin, Cruz, Davies, De Gaudenzi, Dobson, Duesterhoeft, Fazelina, Fosker, Frasch, Fraser, Fuchs, Gabel, Goble, Goffeau, Harris, Hertz-Fowler, Hilbert, Horn, Huang, Klages, Knights, Kube, Larke, Litvin, Lord, Louie, Marra, Masuy, Matthews, Michaeli, Mottram, Muller-Auer, Munden, Nelson, Norbertczak, Oliver, O'Neil, Pentony, Pohl, Price, Purnelle, Quail, Rabbinowitsch, Reinhardt, Rieger, Rinta, Robben, Robertson, Ruiz, Rutter, Saunders, Schafer, Schein, Schwartz, Seeger, Seyler, Sharp, Shin, Sivam, Squares, Squares, Tosato, Vogt, Volckaert, Wambutt, Warren, Wedler, Woodward, Zhou, Zimmermann, Smith, Blackwell, Stuart, Barrell and Myler2005), and 2 years later, the first analysis of catalytic domains of protein phosphatases in Trypanosomatids was also reported. The authors identified 86 phosphatases in Trypanosoma cruzi, 78 in Trypanosoma brucei and 88 in L. major. In the case of L. major, 58 serine/threonine phosphatases were reported, of which 30 were from the PPP family, 13 were phosphatases that dephosphorylate the carboxy-terminal domain of RNA polymerase II (F-cell production eukaryotic-like phosphatases) and 15 were PP2C eukaryotic-like phosphatases (Brenchley et al. Reference Brenchley, Tariq, McElhinney, Szoor, Huxley-Jones, Stevens, Matthews and Tabernero2007). Currently, the function of each of the 15 different PP2C from L. major is unknown.
In the present work, we cloned PP2C from L. major and generated a recombinant His-Tag PP2C fusion protein. LmPP2C was expressed in BL21 E. coli cells and purified. The biochemical characterization of LmPP2C showed it had a molecular mass of 44·9 kDa, an activity optimum at alkaline pH, that it could specifically dephosphorylate a threonine substrate and had a dependence on divalent cations (Mg+2 and Mn+2) for enzymatic activity; all of which are also biochemical characteristics of PP2C from mammalian cells. In all the PP2C, the catalytic domains are important because they include conserved motifs that contain invariant residues involved in the binding of the two metal cations (D42, 66, 202 and 240) and on the phosphate group of the substrate (R37) (Das et al. Reference Das, Helps, Cohen and Barford1996). Alignment of the phosphatase domains of T. gondii with other PP2C proteins showed an absence of two conserved aspartic acid residues that are important for metal binding and typical PP2C activity (Gilbert et al. Reference Gilbert, Ravindran, Turetzky, Boothroyd and Bradley2007). The absence of these residues caused that the recombinant PP2C protein showed low metal-dependent phosphatase activity.
Examination of the primary sequence of LmPP2C shows homology to other PP2C proteins. Additionally, the conservation of the essential catalytic residues between human PP2C (HsPP2C) and LmPP2C suggests that the latter is a PP2C from the PPM family. The analysis of sequences for the PPM phosphatase proteins has identified 11 motifs, which are also conserved between the different phosphatases. The sequence alignment of LmPP2C with the consensus sequences of HsPP2C and LcPP2C phosphatases indicated that LmPP2C contains also 11 conserved motifs characteristic of PP2C. In other microorganisms, such as bacteria, it has been observed that motifs 5a and 5b (Flap domain) of the PP2C protein are absent, thus affecting the enzymatic activity (Schlicker et al. Reference Schlicker, Fokina, Kloft, Grune, Becker, Sheldrick and Forchhammer2008). On the other hand, PP2Cα has a Flap domain, which is inserted between strands β8 and β9 of the catalytic domain (Das et al. Reference Das, Helps, Cohen and Barford1996; Pullen et al. Reference Pullen, Ng, Sung, Good, Smith and Alber2004; Schlicker et al. Reference Schlicker, Fokina, Kloft, Grune, Becker, Sheldrick and Forchhammer2008). This domain, with conserved motifs 5a and 5b, is found in PP2C phosphatases, like the mitochondrial PDP and the phosphatase domains of fungal adenylate cyclases, but is absent in bacterial SpoIIE-type phosphatases (Kennelly, Reference Kennelly2001; Pullen et al. Reference Pullen, Ng, Sung, Good, Smith and Alber2004). The function of the Flap domain is uncertain, but appears to be involved in substrate recognition (Schlicker et al. Reference Schlicker, Fokina, Kloft, Grune, Becker, Sheldrick and Forchhammer2008; Su et al. Reference Su, Schlicker and Forchhammer2011).
The phylogenetic analysis showed that the 15 L. major PP2C are clustered in four groups. Surprisingly, the recombinant protein LmjF.25.0750 and the different species LmxM.25.0750 are grouped in the same cluster suggesting that they share a common ancestor, and, more distant in the same cluster, is LmjF.30.0380. The groups in the phylogenetic tree of PP2C represent proteins with highly similar sequences; however, to the best of our knowledge, there are no reports of their structure, localization in the parasite or functional results.
Using a general substrate for phosphatases, LmPP2C showed a strong preference for Mn+2, in comparison to Mg+2. The preference for this cation has also been observed in other PP2C enzymes like the STP1 phosphatases of Streptococcus agalactiae and Pseudomonas aeuroginosa (Mukhopadhyay et al. Reference Mukhopadhyay, Kapatral, Xu and Chakrabarty1999; Rajagopal et al. Reference Rajagopal, Clancy and Rubens2003). In vitro assays using p-NPP as a substrate showed that the highest specific activity of LmPP2C was at pH 8·5 in the presence of Mg+2; however, when using phosphotreonine, the optimal pH was observed at 7·0. It is well known that PP2C of pathogenic microorganisms show alkaline optimal pH, as has been observed in Prp C from Bacillus subtilis and PPC6803 from Synechocystid (Obuchowski et al. Reference Obuchowski, Madec, Delattre, Boel, Iwanicki, Foulger and Seror2000; Ruppert et al. Reference Ruppert, Irmler, Kloft and Forchhammer2002). For LmPP2C, both divalent cations (Mg+2 and Mn+2) are important for catalytic activity at alkaline pH.
We show novel data on the ultrastructural localization of PP2C in L. major promastigotes, which was found in the flagellum, the flagellar pocket and in the micro axoneme. The heterogeneous staining pattern of the parasites may be due to different development stages of the parasite, since the culture was not synchronized. In other Trypanosomatid parasites, such as Trypanosoma rangeli, a PTP was reported to be associated with the parasite flagellum, yet the function remains unknown (Prestes et al. Reference Prestes, Bayer-Santos, Hermes Stoco, Sincero, Wagner, Umaki, Fragoso, Bordignon, Steindel and Grisard2012). In most of these cells, the PP2C enzymes are mainly localized in the cytosol, and in the case of T. gondii, the parasite also uses secretion from the rhoptries during invasion, in order to deliver a parasite-derived PP2C into the host cell and target it to the host nucleus (Gilbert et al. Reference Gilbert, Ravindran, Turetzky, Boothroyd and Bradley2007).
The localization of PP2C in the flagellum of L. major, an important structure for differentiation in Trypanosomatids, is noteworthy since the phosphatase could potentially exert a regulatory function within this organelle. Yet the role of L. major PP2C remains to be established and functional tests are warranted to determine its biological role in the parasite.
Acknowledgements
This work is one of the requirements to obtain the Ph.D. degree in Posgrado en Ciencias Biológicas (UNAM) for Alma Reyna Escalona Montaño who was the recipient of doctoral fellowship from CONACyT México (fellowship165409, CVU 165409).
The authors gratefully acknowledge Marco Gudiño Zayas, Norma Salaiza Suazo, José Delgado Domínguez, Marco Benítez Rosas, Rocely Cervantes Sarabia, Augusto González Canto, Arturo Wilkins Rodríguez and Angélica Leticia Serrano Ahumada for technical assistance from the Unidad de Investigación en Medicina Experimental, Facultad de Medicina, UNAM. Thanks to Mónica Mondragón, Ricardo Mondragón González and Sirenia González from the Biochemistry Department and Electron Microscopy Facility-LANSE, respectively, at CINVESTAV, México, for the technical support.
Financial support
This work was supported partially by grants 152433 from Consejo Nacional de Ciencia y Tecnología (CONACyT) of México and IN220816 from DGAPA-PAPIIT to MMAG.