Significant outcomes
-
1. The polymorphisms studied were not associated with sialorrhea, but two proxy-SNPs in ERBB4 and TACR1 genes were associated with dry mouth.
-
2. Use of valproate and antipsychotic polypharmacy increased the risk of sialorrhea.
Limitations
-
1. Smaller effects may have been missed because of the small number of genotyped patients compared to a large number of polymorphisms, even after limiting the number of SNPs by minor allele frequency and using proxy-SNPs representing multiple linked SNPs.
-
2. The amount of salivation and dryness of mouth was assessed with quite limited questions.
-
3. Patients who had stopped clozapine medication because of severe sialorrhea may have been selected out of the data because most of the patients had used clozapine for over a year.
Introduction
Clozapine is the most efficient antipsychotic drug for patients with treatment resistant schizophrenia (Lally & Maccabe, Reference Lally and Maccabe2015). It has fewer extrapyramidal adverse effects than previous antipsychotic drugs, but several other adverse effects limit its use. Hypersalivation is one of the most common adverse effects of clozapine. It is experienced by approximately 30 percent of patients, but the percentage varies in the literature. (Praharaj et al., Reference Praharaj, Arora and Gandotra2006) The highest reported prevalence of sialorrhea is 91.8 percent of the clozapine-treated patients (Maher et al., Reference Maher, Cunningham, O’Callaghan, Byrne, Mc Donald, McInerney and Hallahan2016). Sialorrhea is negatively affecting patients’ life. It causes discomfort and psychological complications, for example, embarrassment and low self-esteem.(Liang et al., Reference Liang, Ho, Shen, Lee, Yang and Chiang2010) One severe but rare possible impact of sialorrhea is the aspiration of saliva which can lead to aspiration pneumonia (Bird et al., Reference Bird, Smith and Walton2011). Sialorrhea is among the most common reasons for discontinuing clozapine (Legge et al., Reference Legge, Hamshere, Hayes, Downs, O'donovan, Owen, Walters and Maccabe2016). Dry mouth is also a common adverse effect and it is caused by the anticholinergic properties of clozapine (Kelly et al., Reference Kelly, Conley, Richardson, Tamminga and Carpenter2003). Dry mouth is not known to be among the reasons for discontinuing clozapine, but it might be accompanied with other anticholinergic symptoms such as tachycardia, dizziness, and constipation which are common reasons for discontinuation. (Legge et al., Reference Legge, Hamshere, Hayes, Downs, O'donovan, Owen, Walters and Maccabe2016)
The mechanisms behind clozapine-induced sialorrhea are multifactorial. Clozapine acts as an antagonist or agonist to many neurotransmitters that also play roles in the regulation of salivation. Salivary glands are controlled by the parasympathetic nervous system which releases acetylcholine to bind to muscarinic receptor 3 (CHRM3) and muscarinic receptor 1 (CHRM1) (Tobin et al., Reference Tobin, Giglio and Lundgren2009). Clozapine is an antagonist to CHRM1, muscarinic receptor 2 (CHRM2), CHRM3 and muscarinic receptor 5 (CHRM5) and alfa-2 adrenoreceptor (ADRA2A) but an agonist to muscarinic receptor 4 (CHRM4). Animal studies suggest that clozapine’s antagonism to CHRM1 in acinar cells causes salivary secretion (Ekström et al., Reference Ekström, Godoy and Riva2010). One hypothesis is that ADRA2A antagonists increase salivary output by sympathetic stimulation. (Phillips et al., Reference Phillips, Szabadi and Bradshaw2000). Another hypothesis is that clozapine’s agonism to CHRM4 in salivary glands leads to increase in secretion. (Zorn et al., Reference Zorn, Jones, Ward and Liston1994). Clozapine’s antimuscarinic effects at CHRM2 and CHRM3 may disturb the function of pharyngeal and oesophageal smooth muscles leading to impaired swallowing of saliva. (Praharaj et al., Reference Praharaj, Arora and Gandotra2006) Clozapine also has a high affinity, as an antagonist, for dopamine D4 receptors (DRD4). Dopamine D4 receptors are expressed in the prefrontal cortex and in the nucleus tractus solitarius and nucleus ambiguus which play prominent roles in different phases of swallowing. (Rajagopal et al., Reference Rajagopal, Sundaresan, Rajkumar, Chittybabu, Kuruvilla, Srivastava, Balasubramanian, Jacob and Jacob2014)
Substance P acts as a neurotransmitter in the parasympathetic nervous system and is a neuropeptide known to enhance the swallow response. (Jin et al., Reference Jin, Sekizawa, Fukushima, Morikawa, Nakazawa and Sasaki1994) Substance P bonds to the tachykinin 1 receptor (TACR1) (Severini et al., Reference Severini, Improta, Falconieri-erspamer, Salvadori and Erspamer2002). In salivary glands, there are also epidermal growth factor receptors (ErbB) for epidermal growth factor (EGF). Clozapine affects ErbB kinases which leads to downregulating EGF. (Kobayashi et al., Reference Kobayashi, Iwakura, Sotoyama, Kitayama, Takei, Someya and Nawa2019) EGF has an important role in protecting the oral cavity. (Sarosiek et al., Reference Sarosiek, Bilski, Murty, Slomiany and Slomiany1988)
The activity of G-protein receptor kinases GRK2, GRK3, and GRK5 in cholinergic neurons increases sensitivity to muscarinic receptors (Daigle & Caron, Reference Daigle and Caron2012). There has been found an association between clozapine and expression of G-protein receptor kinases (Ahmed et al., Reference Ahmed, Gurevich, Dalby, Benovic and Gurevich2008). Vasoactive intestinal peptide (VIP) increases salivary gland blood flow and protein secretion and, in rats, evokes fluid secretion. Also, a synergistic interaction between VIP and clozapine has been found. When rats were given a combination of clozapine and VIP, saliva secretion increased more than when given VIP and clozapine separately (Ekström et al., Reference Ekström, Godoy, Loy and Riva2014).
There have been some studies that have formerly found an association between clozapine-induced sialorrhea and genetic polymorphisms. A single nucleotide polymorphism (SNP) in the ADRA2A gene (rs1800544) has been reported to be associated with sialorrhea among patients treated with clozapine. Increased sialorrhea was associated with ADRA2A rs1800544 CC genotype in comparison with G-allele carriers (CG and GG genotypes). (Solismaa et al., Reference Solismaa, Kampman, Seppälä, Viikki, Mäkelä, Mononen, Lehtimäki and Leinonen2014). A 120-base pair duplication in DRD4 has also been reported to be associated with clozapine-induced sialorrhea. (Rajagopal et al., Reference Rajagopal, Sundaresan, Rajkumar, Chittybabu, Kuruvilla, Srivastava, Balasubramanian, Jacob and Jacob2014).
As the genetic background of clozapine-induced sialorrhea is still poorly known, it is necessary to further investigate possible genetic associations with sialorrhea and dry mouth by selecting a broad group of genes (TACR1, VIP, ERBB1, ERBB2, ERBB4, GRK2, GRK5, CHRM1, CHRM3, CHRM4, ADRA2A, and DRD4) related to salivary gland regulation. Proteins coded by selected genes have roles in the regulation of saliva secretion or swallowing or are present in salivary glands as explained above. The study explores the possible genetic mechanisms behind the variation in intensity of sialorrhea and dry mouth among patients treated with clozapine.
Material and methods
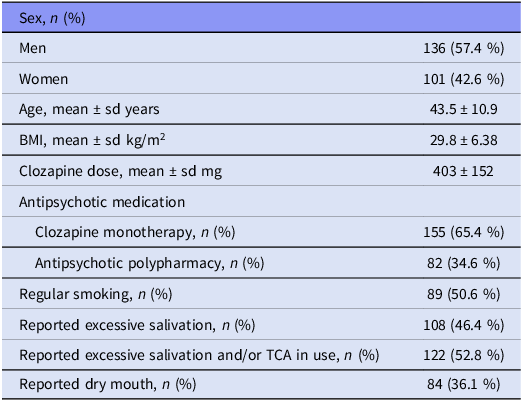
The study population consists of 237 clozapine-treated patients who have diagnosis of schizophrenia, schizoaffective or other nonorganic and non-affective psychoses according to the International Classification of Diseases, 10th Revision (World Health Organization (WHO) 1993). All patients were over 18 years old, Caucasian, of Finnish origin. Written consent was collected from all the patients. The study was conducted in three hospital districts in Finland (Satakunta, Pirkanmaa and Southern Ostrobothnia). The Ethics Committee of Satakunta Hospital District approved the study. Characteristics of the patients are presented in Table 1.
Table 1. Demographics and clinical characteristics of the study participants
Fasted blood samples for concentration measurements were collected in the morning, following a 12-hour interval post the evening dose. All patients were on a stable dosage of clozapine. The concentrations of clozapine and norclozapine in serum were quantified through the application of liquid chromatography in a commercial laboratory.
Measuring of salivation was carried out by using the Liverpool University Neuroleptic Side Effect Scale (LUNSERS) (Day et al., Reference Day, Wood, Dewey and Bentall1995) where patients rate the severity of 51 different side effects from 0 to 4 (0 = not at all, 1 = very little, 2 = a little, 3 = quite a lot and 4 = very much).
The salivation was analysed in three ways. First, the answer of LUNSERS symptom number 37 (over-wet drooling mouth) was divided into two different variables: ‘excessive salivation’ and ‘non-excessive salivation’. Secondly, patients using tricyclic antidepressants (TCA) for excessive salivation were added to the ‘excessive salivation’ group even if they did not report sialorrhea in the questionnaire. Of the 32 patients using TCA, two were excluded because their indication for this medication was not sialorrhea.
LUNSERS symptom number 6 (dry mouth) was also divided into two groups (patients who did not have dryness of the mouth at all (0) vs. those who had from very little to very much dryness of mouth (1–4)) and analysed as a dichotomous variable. Number and percentage of patients reporting excessive salivation or dry mouth are presented in Table 1.
Genotyping and imputation
Genomic DNA was extracted from peripheral blood leukocytes using a QIAamp DNA Blood Midikit and an automated biorobot M48 extraction (Qiagen, Hilden, Germany). Samples were genotyped using an Illumina Infinium HumanCoreExome-12 DNA Analysis Beadchip, version 1.0., according to the manufacturer’s instructions at Helmholtz Zentrum, M FC;nchen, Germany. The following quality control filters were applied: GenCall score < 0.15, GenTrain score less than 0.20, sample and an SNP call rate less than 0.95, Hardy-Weinberg equation P value less than × 10−6, excess heterozygosity, cryptic relatedness (pi-hat > 0.2), gender check, and multidimensional scaling. After quality control, genotype samples and 531,983 SNPs were available for 172 patients.
Imputation was performed in two stages: haplotype phasing was done using SHAPEIT v2 and genotype imputation using IMPUTE2 v. 2.3.2 and 1000 Genomes Phase I integrated variant set haplotypes as a reference. SNPs with info ≥ 0.3 were considered well imputed.
A total of 9039 polymorphisms (SNPs including insertions and deletions) from the selected genes (17 ADRA2A, 32 CHRM1, 1592 CHRM3, 6 CHRM4, 36 DRD4, 580 ERBB1, 57 ERBB2, 4879 ERBB4, 21 GRK2, 901 GRK5, 365 IP3, 463 NK1, 73 NTS, and 17 VIP) were available after genotyping and imputation for analyses.
Statistical methods
To assess the distribution of continuous variables, Kolmogorov-Smirnov test was applied. Exploratory two-variable comparisons between sialorrhea or dry mouth and age, sex, BMI, smoking, clozapine and norclozapine concentrations, clozapine/norclozapine ratio, use of other medications (antidepressants, other antipsychotics, mood stabilisers, benzodiazepines, and blood pressure medication) were analysed with t-test, or Mann-Whitney U test if the variable was not normally distributed, and Pearson’s chi-squared test. For the analyses, all antipsychotic doses were converted to chlorpromazine equivalents (Aronson, Reference Aronson2009).
Genetic associations were analysed with multiple linear and logistic regression models explaining sialorrhea and dry mouth with each SNP tested separately in the model as coefficients and including other coefficients selected after the exploratory analyses or otherwise hypothesised to be linked with sialorrhea or dry mouth. P values were adjusted using the false discovery rate (FDR) method for multiple testing correction. Due to the high number of SNPs related to the number of patients genotyped, a clumping of SNPs was performed by using one proxy SNP for SNPs that had a linkage disequilibrium of r2 > 0.2 with a window of one million base pairs (500k base pairs downstream and 500k pairs upstream). Selection of the proxy SNP was made favouring the lowest p-value in the logistic and linear regression models and the lowest minor allele frequency (MAF). SNPs with a MAF below 0.1 were excluded. Best fitting models were selected. The combined effect of multiple proxySNPs with an FDR-adjusted p < 0.05 was analysed by forming a genetic risk score by multiplying the dosage of the SNP by the estimate in the previous regression models and furthermore summing the estimate-weighted dosages of the SNPs.
Results
Age, sex, regular smoking, and BMI were not associated with sialorrhea or dryness of mouth in clozapine-treated patients.
Potential factors relating to the mouth dryness
Clozapine dose, clozapine or norclozapine concentration and clozapine to norclozapine ratio were not associated with dryness of mouth. There was a trend of dose of selective serotine reuptake inhibitor medication being associated with dryness of the mouth: mean doses were 8.17 ± 16.0 fluoxetine equivalent mg in patients who did not report dryness of mouth vs 13.4 ± 20.3 mg fluoxetine equivalent mg in those who did (Mann-Whitney U = 4055, p = 0.055).
Potential factors relating to the sialorrhea
Clozapine dose, clozapine or norclozapine concentration and clozapine to norclozapine ratio were not associated with excessive salivation.
Chi square test statistics of comparisons between use of different additional medications and excessive salivation are presented in Table 2. Patients using mood stabilisers suffered from excessive salivation more commonly than the group that did not use mood stabilisers. In more detailed comparisons, the association seems to be related particularly to patients on valproate medication reporting excessive salivation whereas other mood stabilisers were not associated with sialorrhea. Patients who used other antipsychotics in addition to clozapine also had increased salivation. The higher dose of other antipsychotic drug was associated with excessive salivation (Mann-Whitney U = 5157, p = 0.009). Doses of antipsychotics other than clozapine are presented in Figure 1 in non-excessive and excessive salivation groups. There was a trend of benzodiazepine users having more salivation compared to those who do not use benzodiazepines when the use of tricyclic antidepressants was included in the ‘excessive salivation’ variable. Beta-blockers and blood pressure medication were not associated with excessive salivation or dryness of the mouth.
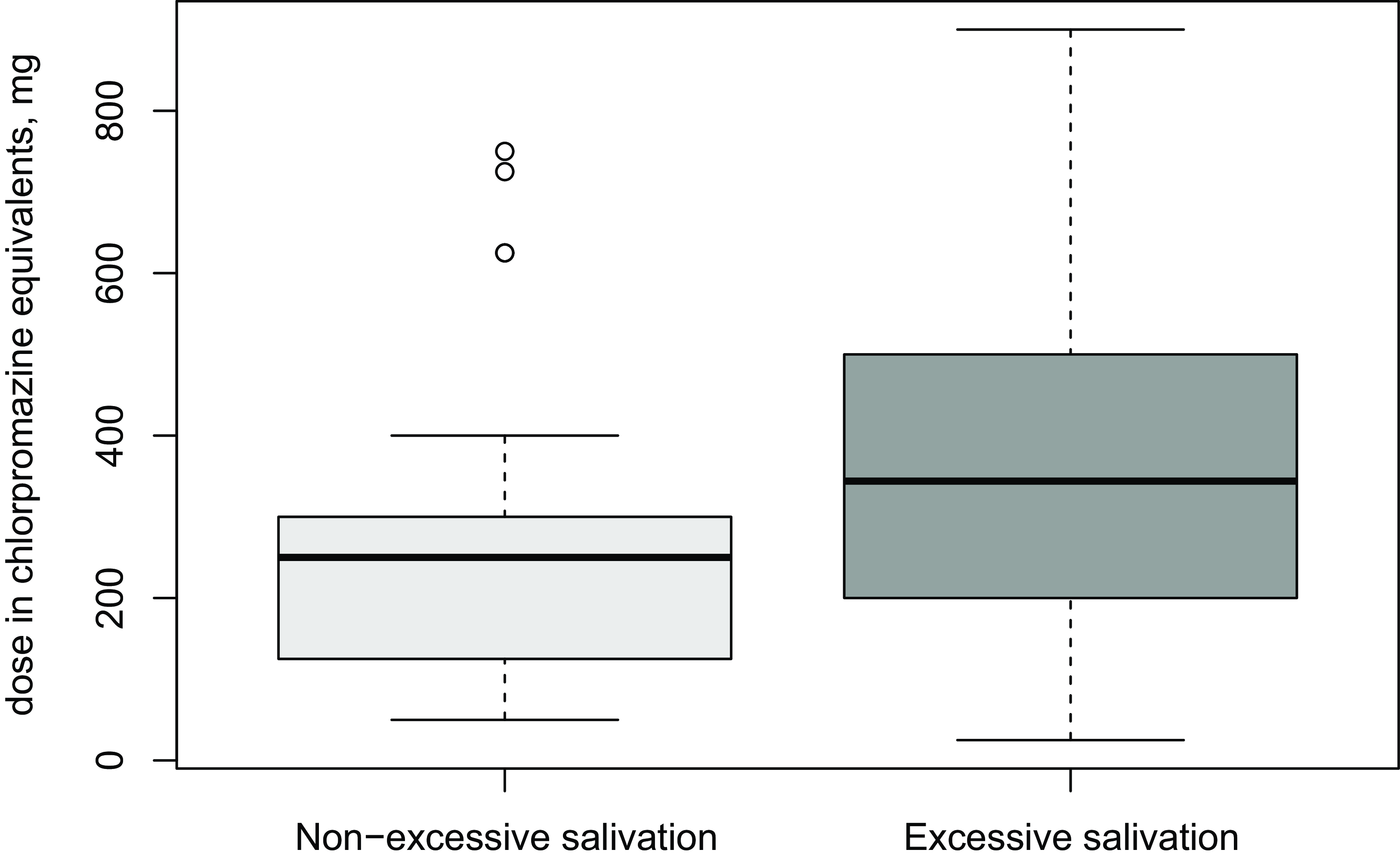
Figure 1. Doses of other antipsychotic drugs than clozapine in two groups: non-excessive salivation and excessive salivation.
Table 2. Comparisons between salivation groups (no excessive salivation vs. excessive salivation) and different other medications used in addition to clozapine
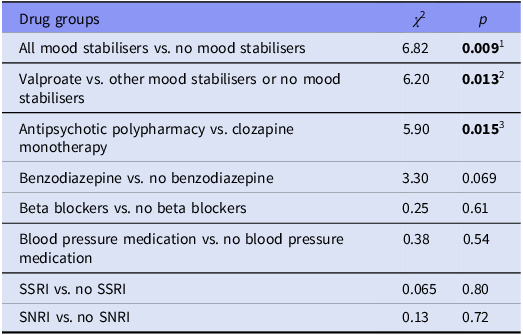
1 Patients on mood stabilisers had a higher risk of excessive salivation than those not on mood stabilisers.
2 Valproate users were more likely to experience excessive salivation compared to those on other mood stabilisers or none.
3 Antipsychotic polypharmacy increased the risk of excessive salivation compared to using clozapine alone.
Potential roles of genetic variations in the development of dry mouth following clozapine therapy
After clumping, the number of polymorphisms decreased from 9039 to a total of 97 proxy-SNPs. In linear and logistic regression analyses explaining sialorrhea with various coefficients and the proxy-SNPs, no proxy SNP was associated with sialorrhea. In analyses explaining dry mouth with logistic regression with age and sex as coefficients, two proxy-SNPs were associated with dry mouth: ERBB4 rs3942465 (FDR-adjusted p = 0.025) and TACR1 rs58933792 (FDR-adjusted p = 0.029). A combined effect of these SNPs in a logistic regression model is presented in Table 3. When regular smoking was added to the model, the SNPs associations were not statistically significant, a trend of association remained for ERBB4 rs3942465 (FDR-adjusted p = 0.051), although regular smoking was not associated with dry mouth. SNPs for which ERBB4 rs3942564 and TACR1 rs58933792 were used as proxies and had an r2 over 0.2 are presented in Supplementary Table 1.
Table 3. Logistic regression model explaining dry mouth (no dryness of mouth vs. at least very little dryness of the mouth) with the following coefficients: age, sex, and score formed of ERBB4 rs3942465 and TACR1 rs58933792 SNPs
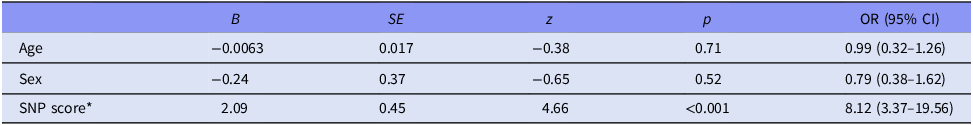
* Estimate-weighted genetic risk score of two SNPs: ERBB4 rs3942465 and TACR1 rs58933792.
Discussion
The focus of this study was to find possible associations with sialorrhea, dry mouth and the studied polymorphisms in the selected genes related to the regulation of salivation. No associations between sialorrhea and studied polymorphisms were found. Clozapine dose, clozapine or norclozapine concentration and clozapine to norclozapine ratio did were not associated with sialorrhea or dryness of mouth. The use of valproate or additional antipsychotic medication with clozapine was associated with increased sialorrhea.
Proxy-SNPs in ERBB4 (rs3942465) and TACR1 (rs58933792) were associated with dry mouth. ERBB4 (rs3942465) was a proxy SNP for 676 SNPs in ERBB4 and TACR1 rs58933792 for 153 SNPs. Both proxy-SNPs are in the intron region of their respective genes. No highly correlated (r2 > 0.8) SNPs were in linkage disequilibrium with the proxy-SNPs. Two of the lesser correlated (r2 < 0.8) SNPs in TACR1 (rs17010664 and rs72824221) are located in 3’-UTR and the rest are located in the intron region. Also, one deletion (chr2.213404504) in the promoter region of ERBB4 was linked to the proxy SNP. There are no previous publications of the above-mentioned SNPs or the deletion. In this study, the TACR1 gene was studied because clozapine affects the parasympathetic nervous system and substance P is a neurotransmitter in the parasympathetic nervous system. The hypothesis is that activity of substance P could cause sialorrhea by reducing swallow response and causing accumulation of saliva. Dysphagia is known as a rare but severe adverse effect of clozapine (Crouse et al., Reference Crouse, Alastanos, Bozymski and Toscano2018). ERRB4 is receptor-tyrosine-receptor kinase. Erbb4 kinases might be molecular targets of clozapine (Kobayashi et al., Reference Kobayashi, Iwakura, Sotoyama, Kitayama, Takei, Someya and Nawa2019). EGF has an important role in protecting the oral cavity and there are EGF receptors in salivary glands. Nevertheless, it is still unclear what is the role of EGF in the regulation of saliva, thus the mechanism by which the deletion in ERBB4 would cause dryness of mouth remains unknown. Formatting…
Clozapine has both anticholinergic and cholinergic effects. This explains why clozapine can cause both dry mouth and sialorrhea. (Ignjatović Ristić et al., Reference Ignjatović Ristić, Cohen, Obradović, Nikić-Đuričić, Drašković and Hinić2018) The percentage of patients on clozapine medication having dry mouth varies from 0% (Tollefson et al., Reference Tollefson, Birkett, Kiesler and Wood2001) to 20% (Kelly et al., Reference Kelly, Conley, Richardson, Tamminga and Carpenter2003). Previously it has been found that patients with high clozapine concentration in plasma have more sialorrhea (Schoretsanitis et al., Reference Schoretsanitis, Kuzin, Kane, Hiemke, Paulzen and Haen2021) and it is commonly managed by lowering the dose (Stroup & Gray, Reference Stroup and Gray2018). In this study, surprisingly, a similar association was not found; sialorrhea occurred regardless of the dosage and concentrations. Clozapine users who also used mood stabilisers suffered more from excessive salivation. Previously it has been found that lithium, one of the mood stabilisers, causes both sialorrhea and dry mouth. (Alexopoulos et al., Reference Alexopoulos, Streim, Carpenter and Docherty2004) The higher dose of other antipsychotic drugs, in patients who use other antipsychotics in addition to clozapine, was associated with excessive salivation. The reason for this is unclear because sialorrhea is generally known to be associated only with clozapine use and not with other antipsychotics (Stroup & gray, Reference Stroup and Gray2018).
The strength of the study is that salivation-related genes and their associations with clozapine-induced sialorrhea have not been examined as broadly formerly. This is also the first study suggesting that genotypes can affect the dryness of the mouth during clozapine treatment. The SNP in the ADRA2A gene (rs1800544) that was previously found to be associated with sialorrhea was not associated with sialorrhea in this study (Solismaa et al., Reference Solismaa, Kampman, Seppälä, Viikki, Mäkelä, Mononen, Lehtimäki and Leinonen2014). . Limitations of the study include that the sample size was limited and quite restrictive in relation to the number of polymorphisms. It is possible that smaller effects were missed because of the data’s small size when examining larger numbers of genes even though the number of polymorphisms was reduced with MAF-limitation and clumping. This is possibly the reason that the single SNP ADRA2A rs1800544 was not associated with sialorrhea in this study. Another limitation is that the in-depth mechanism of found proxy-SNPs and effect on for example mRNA expression was not assessed. Another limitation is that the amount of salivation was asked with only two questions in LUNSERS. Most of the examined patients had used clozapine for over a year. Therefore, some patients who had to stop the medication early because of difficult sialorrhea, have probably been selected out of the data.
In conclusion, this study suggests that genetic variations in ERBB4 and TACR1 genes contribute to experienced dryness of mouth in clozapine-treated patients. Use of valproate and antipsychotic polypharmacy was associated with sialorrhea, but dose or concentration of clozapine was not.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/neu.2024.9.
Author contributions
Olli Kampman and Niko Seppälä contributed to planning and designing the original study and data collection. Niko Seppälä and Merja Viikki participated locally in acquisition of data. Hanna Puolakka, Anssi Solismaa and Olli Kampman were responsible for planning the present study and forming the present research hypothesis. Leo-Pekka Lyytikäinen, Nina Mononen and Terho Lehtimäki participated in planning and performing the laboratory analyses and genotyping and provided the methodology for laboratory analyses. Hanna Puolakka, Anssi Solismaa and Leo-Pekka Lyytikäinen conducted the statistical analyses. Hanna Puolakka, Anssi Solismaa, and Olli Kampman contributed to the interpretation of the data. Hanna Puolakka, Anssi Solismaa and Olli Kampman drafted the article. Olli Kampman, Merja Viikki, Nina Mononen, Niko Seppälä, Terho Lehtimäki, Leo-Pekka Lyytikäinen revised the article. All authors have reviewed and approved the article.
Financial support
Supported by the Competitive Research Funding of the Tampere University Hospital, Signe and Ane Gyllenberg Foundation, EU Horizon 2020 (grant 848146 for To Aition); Tampere University Hospital Supporting Foundation, and Finnish Society of Clinical Chemistry.
Competing interests
Niko Seppälä has given lectures and worked as a Clinical Advisor at Viatris, Finland. Niko Seppälä is an Advisory Board Member at Viatris, Italy.
Hanna Puolakka, Anssi Solismaa, Leo-Pekka Lyytikäinen, Merja Viikki, Nina Mononen, Terho Lehtimäki and Olli Kampman declare no competing interests.
Ethical standard
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008. The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.







