INTRODUCTION
Rift Valley fever virus (RVFV) is an arthropod-borne virus (family Bunyaviridae, genus Phlebovirus). Its single-stranded RNA genome is tri-segmented and encodes two glycoproteins, a viral polymerase, a nucleoprotein and two non-structural proteins. RVF is a zoonosis of major concern in Africa and the Arabian Peninsula. It can be transmitted by a broad range of mosquitoes and has been isolated from more than 30 species of six genera [1, Reference Fontenille2].
RVF was first described in 1931 in sheep, cattle and humans in Kenya [Reference Daubney3]. Clinical manifestations can be multifold, depending on animal age and species. The infection is most significant in productive livestock such as small ruminants, cattle and camels. Characteristic ‘abortion storms’ with up to 100% newborn case-fatality rates, can be seen during acute infections. Multiple organ infection and necrosis can be found in aborted and malformed infected fetuses. In adult animals a severe progression, often accompanied by hepatic failure, is mainly reported for small ruminants. However, only moderate clinical signs are observed in infected adult cattle and camels. Most human infections proceed as mild flu-like illnesses. In some cases encephalitis, retinitis, necrotizing hepatitis or even haemorrhagic fever syndromes are observed [Reference Gerdes4].
After the initial discovery of RVFV, larger epidemics were reported from other African countries, such as South Africa, Tanzania, Zambia, Kenya, Zimbabwe, Egypt and Madagascar [Reference Gerdes4]. A considerable outbreak in Saudi Arabia and Yemen in 2000 was caused by trade and movement of infected ruminants [Reference Balkhy and Memish5]. Besides animal losses, RVFV epidemics incur substantial financial and socio-economic costs for agriculture, the labour market and healthcare [Reference Rich and Wanyoike6].
In Mauritania the first known RVF epidemic in 1987, causing 220 human deaths, was mainly associated with the construction of the Diama Dam retaining the Senegal River [Reference Saluzzo7–Reference Jouan10]. Since then outbreaks have occurred periodically in 1993 [Reference Zeller, Akakpo and Ba11], 1998 [Reference Nabeth12] and 2003 [Reference Faye13]. The epidemic reported in 2010 [Reference El Mamy14, Reference Jäckel15] emphasized the role of camelids, as for the first time severe clinical manifestations were observed in this species. In 2012 unusual heavy rainfalls led to a further outbreak of RVF in southern Mauritania. Abortions in ruminants and haemorrhagic fever symptoms in humans with 13 fatal cases were reported, mainly occurring in the Mauritanian regions of Tagant, Brakna, Trarza, Assaba and Hodh El Gharbi [Reference Sow16]. After an inter-epidemic period, a further outbreak occurred in September 2013 affecting sheep, goats and camels. Foci of this outbreak were again seen in Trarza and Brakna, close to the Senegalese border [17]. Finally, from September 2015 to March 2016 a RVF outbreak occurred, affecting sheep and goats [18]. A markedly reduced duration of inter-epidemic periods was observed considering past outbreaks of RVFV in Mauritania. Retrospective investigations revealed that the likelihood of extensive outbreaks mainly depends on precipitation and presence of competent vectors [Reference Caminade19]. In periods with low-level precipitation, the virus is presumably maintained by infrequent transmission from vertically infected Aedes spp. to susceptible intermediate hosts, such as wildlife or livestock [Reference Diallo20, Reference Pepin21]. However, these intermediate hosts have not yet been fully described.
To date, little is known about ongoing RVFV infections in endemic regions during inter-epidemic periods. To elucidate the potential of enzootic infections and to compare the immunological status between epidemic and inter-epidemic periods, it is necessary to examine prevalence and potential infections in susceptible hosts during these periods. We analysed 1066 samples of sheep, goats, cattle and camels that were collected in inter-epidemic periods (January–March 2012, January–June 2013) in Mauritania for RVFV infections, using a systematic multi-stage serological and molecular analysis. The analysis encompassed enzyme-linked immunosorbent assays (ELISA), indirect immunofluorescence assays (IIFA), serum neutralization test (SNT) as well as quantitative real-time RT–PCR (qRT–PCR).
Considering the specific period of sampling between different outbreaks, these data will provide insights into variations and characteristics of antibodies in livestock and potential carriers of RVFV during inter-epidemic periods. The obtained results can form the basis for future surveillance and monitoring in Mauritania.
MATERIALS AND METHODS
Sampling and treatment of samples
In total, 1066 animals of productive livestock were sampled in Mauritania in 2012 and 2013 under the direction of the Centre National de l'Elevage et de Recherches Veterinaires (CNERV). In detail, blood samples of 497 small ruminants (294 goats, 158 sheep, 45 undetermined), 488 cattle and 81 camels were collected. Species, sampling location, preliminary reports of abortions and, if possible, age of animals were recorded. Sampling of both small ruminants and camels included regions of known epidemic areas [Reference Faye13–Reference Jäckel15]. As there is no data about apparent RVFV infection in cattle in Mauritania, areas of known cattle husbandry were chosen for sampling. Collectively the regions Adrar, Assaba, Brakna, Hodh El Chargui, Hodh El Gharbi, Gorgol, Gouidimak, Inchiri, Nouakchott, Tagant, and Trarza were included (Fig. 1). Blood was drawn by trained personnel through puncture of the vena jugularis according to good veterinary practice. In accordance with safety protocols all sera were inactivated before handling (gamma radiation 30 kGy, Synergy Health, Germany). Additionally, all sera from small ruminants were pretreated by heating at 56 °C for 1 h as described previously [Reference van Vuren and Paweska22].

Fig. 1. Sampling locations. Map of Mauritania indicating sampling regions and sampled species. Both sample sizes and proportions of sampled species (Nouackchott, Inchiri) are indicated by circular charts.
Serological and molecular investigation
Initially, all serum samples were screened with ID Vet competitive ELISA (cELISA) (small ruminants, cattle, camels), which detects antibodies against the nucleocapsid protein (NP). In addition, sera of small ruminants were analysed with the indirect IgG glycoprotein (∆Gn)-based ELISA, allowing the independent comparison of immunoreactivity against the two different antigens. Positive results were confirmed with the SNT, known as the gold standard for serological diagnosis of RVFV infection. In case of a negative SNT, samples were additionally tested by IIFA. Confirmatory results were obtained with SNT and IIFA. Seroprevalence and 95% confidence intervals (CIs) were calculated with R version 2·14·0 (R Foundation, Austria). Test for significance and logistic or linear regression were performed using SAS Enterprise Guide 7.1 (SAS Institute Inc., USA). For both Fisher's exact test and logistic or linear regression a value of P ⩽ 0·05 was considered significant.
For detection of IgM antibodies, all sera which were positive by ID Vet cELISA were tested with the ID Vet IgM capture ELISA (small ruminants and cattle) or the indirect IgM in-house ELISA for camelids. IgM-positive samples were further tested for the presence of viral RNA.
Indirect IgG ∆Gn ELISA
All sera from sheep and goats were tested with the indirect IgG ∆Gn ELISA as described previously [Reference Jackel23]. Briefly, ELISA plates (Maxisorp, Denmark) were coated with 2 µg/ml recombinant ∆Gn protein. After washing, plates were blocked with 10% skim milk. Sera were diluted 1:25 in 2% skim milk. Serum from an immunized rabbit was used as positive control (dilution 1: 20 000). Serum of a German sheep from quarantine facilities of Friedrich-Loeffler-Institut was used as negative control (dilution 1:25). In the next step a 1:5000 protein G dilution was added. Finally 2,2′-azino di-ethylbenzothiazoline sulphonic acid (ABTS, Roche, Germany) was added and after 30 min the reaction was stopped with 1% sodiumdodecyl-sulfate. The plates were read at 405 nm. All samples with a percentage of positive control serum (OD value sample/median of positive control × 100) higher than 20·75 were identified as positive.
cELISA
All serum samples were tested with the ID Screen® RVFV competitive multi-species ELISA (ID Vet, France) according to the manufacturer's instructions. As competitive reactions are detected, both IgG and IgM are indistinguishably identified. The nucleoprotein is used as capture antigen.
IgM capture ELISA
Sera were tested with the ID Screen® Rift Valley fever IgM capture ELISA (ID Vet) for the specific presence of IgM. Because a ruminant-specific anti-IgM antibody is applied, the ELISA is only adaptable for small ruminants and cattle.
SNT
The SNT was performed as described in the OIE Terrestrial Manual [24]. Briefly, 25 µl of 100 TCID50 of RVFV (MP-12 vaccine strain) were added to 25 µl of serial twofold-diluted and heat-inactivated sera. The 96-well microtitre plates were incubated for 30 min at 37 °C in an atmosphere of 5% CO2. Next, 50 µl 3 × 105 Vero 76 cells (Collection of Cell Lines in Veterinary Medicine, Friedrich-Loeffler-Institut, Germany), diluted in minimum essential medium with penicillin, streptomycin and 5% fetal calf serum, were added to each well. Plates were incubated at 37 °C in 5% CO2 for 6 days. Neutralizing doses of 50% (ND50) were expressed as the reciprocal of the serum dilution that still inhibited >50% of cytopathic effect. A serum sample was considered positive with a ND50 of ⩾10. Positive and negative control sera, as well as cell controls were always included. Additionally the TCID50 of the challenge virus was checked via titration in each run. In regular testing procedure, sera were diluted from 1:10 to 1:80. For all samples with ND50 ⩾120 the endpoint ND50 was evaluated with an additional SNT with serum dilutions from 1:40 to 1: 20 480.
IIFA
Sera were tested with a commercial kit for Rift Valley fever virus indirect immunofluorescence (Euroimmun, Germany) with adaptations as described previously [Reference Jäckel15]. Serum samples were used in dilutions of 1:100. The detection of antibodies was achieved with species-specific secondary antibodies. Thus, Cy3-labelled donkey anti-sheep, donkey anti-goat or goat anti-bovine antibodies (Dianova, Germany) were used in a 1:200 dilution. For sera of camels, a polyclonal rabbit anti-camel antiserum (Bethyl Laboratories, USA) in a 1:100 dilution was detected by a Cy3-labelled goat anti-rabbit antibody (Dianova) in a 1:800 dilution. Camel-derived IgM antibodies were detected by goat anti-camel IgM antibody (Triple J Farms, USA, 1:50 dilution) and the Cy3-labelled rabbit anti-goat antibody (1:800 dilution, Dianova). Species-specific positive and negative controls were included in each run.
Indirect IgM in-house ELISA for camelids
ELISA plates (Maxisorp) were coated with 100 µl of 4 µg/ml of recombinant NP in 0·05 m carbonate-bicarbonate buffer overnight at 4 °C. Every second well was coated with 0·05 m carbonate-bicarbonate buffer only. Plates were washed three times with 300 µl phosphate-buffered saline and 0·1% Tween-20 (PBS-T). Blocking was performed with 200 µl of 10% skim milk for 1 h at 37 °C. Sera were diluted 1:25 in duplicate in 2% skim milk. After washing as described previously, 100 µl of each sample were added to both a well with NP and with only carbonate-bicarbonate buffer. Plates were incubated for 1 h at 37 °C and washed with PBS-T. A goat anti-camelid IgM antibody was diluted 1:1000 in 2% skim milk and 100 µl were added to each well and were incubated for 1 h at 37 °C. After washing with PBS-T, a secondary rabbit anti-goat antibody conjugated with horseradish-peroxidase (Dianova) diluted 1:5000 in 2% skim milk was added and incubated for 1 h at 37 °C. Following washing, 100 µl ABTS were added and incubated for 30 min at room temperature in the dark. The reaction was stopped with 1% sodiumdodecyl-sulfate and read at 405 nm.
For final analysis a corrected OD405 was determined by subtracting the OD value of the well without antigen from the OD405 of the well coated with antigen (ΔOD405).
Real-time reverse transcriptase (RT)–PCR and recovery of viral sequences
RNA extraction was performed using the QIAmp® Viral RNA Mini kit (Qiagen, Germany) according to the manufacturer's instructions. Samples were tested with a qRT–PCR targeting the L segment at nucleotide position 2912–3001 [Reference Bird25] using the QuantiTect Probe RT-PCR kit (Qiagen). For each reaction 5 µl RNA, 10 pmol of both forward and reverse primers and 1·25 pmol of the probe were applied in a total volume of 25 µl. PCR reaction conditions were used as follows: 50 °C for 30 min, 95 °C for 15 min and 45 cycles at 95 °C for 10 s, 55 °C for 25 s and 72 °C for 25 s. For quantification a synthetic RNA control was used as described previously [Reference Jäckel15]. Samples with ⩾5 copies/μl RNA were classified as positive.
Partial viral sequences were generated by using the Superscript III One-Step RT–PCR kit (Invitrogen, USA) and primers MRV1a and MRV2 g, targeting a 809 bp region of the M segment [Reference Faye13]. PCR reaction conditions were used as follows: 15 min at 45 °C, 3 min at 95 °C, 40 cycles at 95 °C for 20 s, 55 °C for 30 s, 72 °C for 60 s and finally 7 min at 72 °C. After electrophoretic separation, PCR products were purified using the QIAquick® Gel Extraction kit (Qiagen) as suggested by the manufacturer. The purified PCR products were sequenced (Eurofins Genomics, Germany). Additionally a real-time PCR for the detection of phleboviruses, generating an ~370 bp amplicon of the S segment, was applied [Reference Lambert and Lanciotti26]. PCR products were sequenced (Eurofins Genomics).
Alignment and phylogenetic analysis were performed using BLAST (https://blast.ncbi.nlm.nih.gov/) and Geneious (www.geneious.com) software.
To prevent cross-contamination during RNA isolation and RT–PCR procedures, mechanical barriers (spatial separation, unidirectional traffic, separate instruments) and decontamination procedures were applied. In addition, handling of infected cell cultures was performed in a separate building.
RESULTS
Small ruminant sera (n = 497) were tested with both the commercial cELISA and the indirect IgG ∆Gn ELISA yielding 18 and 29 positive sera, respectively (Table 1). Positive samples were subjected to SNT, where 18 samples with neutralizing activity were found. Neutralizing doses ranged from 10 up to 2560 (Table 2). SNT-negative sera were finally checked by IIFA, which led to the detection of one additional seropositive sample. In summary 19/497 sera (3·8%, 95% CI 2·32–5·91) were found to be seropositive. Comparatively 9·5% (95% CI 5·4–15·17) of sampled sheep and only 1·4% (95% CI 0·37–3·44) of goats were found to be carriers of RVFV-specific antibodies. Samples were obtained from six different collection sites, located in the central and southern regions (Fig. 1). Positive sera were found only in Gorgol (2·6%) and Tagant (11·3%) with significant differences between these two regions (Fisher's exact test, P = 0·036). In other regions small ruminants were free of RVFV-specific antibodies (Table 3). Subsequent testing for IgM antibodies using a commercial IgM capture ELISA for those samples that were positive or inconclusive by ID Vet cELISA revealed no positive cases.
Table 1. Rift Valley fever virus serological results

IIFA, Indirect immunofluorescence assay; SNT, serum neutralization test.
* Confirmation test for ELISA-positive samples only.
† Confirmation test for SNT-negative samples only.
Table 2. Inter-species comparison of neutralizing titres
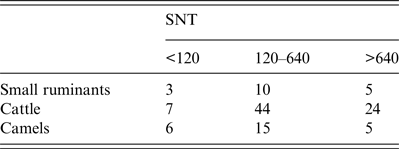
Endpoints of neutralizing doses of 50% (ND50) were determined in serum neutralization test (SNT). For comparison ND50 values were classified as <120, 120–640 or >640.
Table 3. Regional prevalence in small ruminants, cattle and camels

Pos., Positive; Prev., prevalence; CI, confidence interval.
Serological status of positive samples was confirmed in ELISA and serum neutralization test or in ELISA and indirect immunofluorescence. Only if both methods confirmed the presence of RVFV-specific antibodies, samples were classified as positive.
Cattle sera (n = 488) were obtained from six collection sites mainly located in the southern part of Mauritania, near the Senegalese border (Fig. 1). They were first screened with the ID Vet cELISA, encompassing 73 positive and four inconclusive samples. After verification with SNT and IIFA a total of 75 samples were confirmed as positive with an overall prevalence of 15·4% (95% CI 12·28–18·88) (Table 1). ND50 values of SNT-positive sera ranged from 10 to 10 240 (Table 2) displaying the highest neutralizing activity compared to other investigated species. Positive cattle sera were found in all regions with prevalences ranging from 10·6% to 29·4%, without significant regional differences (Fisher's exact test, P = 0·091) (Table 3). A total of 77 cattle samples, which were positive or inconclusive by cELISA, were eventually assayed for the presence of RVFV-specific IgM antibodies using a commercial IgM capture ELISA. Only one bovine sample, derived from a slaughterhouse from Nouakchott (Fig. 1), was detected as positive, which had tested negative by SNT and IIFA. Results were complemented by positive qRT–PCR, which displayed 287 copies/μl RNA (corresponding Ct value 33·64). The results were confirmed by the recovery of partial sequences of the S segment (accession no. KX503062) and M segment (accession no. KX503063). However, due to the short sequences of about 370 nt and 405 nt, respectively, a sound phylogenetic analysis was not possible. A comparison with representative RFVF strains is depicted in Supplementary Figure S1. Due to the previous irradiation of sera, no isolation of virus for comprehensive phylogenetic analysis was possible.
Finally, 81 camel sera were collected in two different regions in central Mauritania (Fig. 1). Twenty-four sera tested positive and two sera inconclusive using the ID Vet cELISA. All 26 reactive samples were verified by SNT yielding ND50 values in the range of 15 to 1920 (Tables 1 and 2). The overall antibody prevalence in camelids was 32·0% (95% CI 22·15- 43·4). Positive sera were found at both collection sides, with a significantly higher prevalence in Adrar compared to Inchiri (Fisher's exact test, P < 0·0001) (Table 3). Since no commercial ELISA for the detection of camelid IgM antibodies is available, the sera were analysed with a newly developed indirect IgM in-house ELISA. All 81 tested samples harboured ΔOD values <0·13 while the positive control ΔΟD value was 0·78. This result is indicative of the absence of IgM antibodies in tested camels. An IgM-positive serum of a camel from Mauritania collected during a RVF outbreak in 2010 [Reference Jäckel15] served as positive control. Determination of IgM antibodies of the positive control had been verified by IIFA. Sera of an alpaca from quarantine facilities of Friedrich-Loeffler-Institut and a camel from a German zoo were used as negative controls (Supplementary Fig. S2).
Correlations between age of animals and presence of RVFV-specific antibodies were calculated for all animals with recorded ages. Further information regarding age of animals was available for 354/497 small ruminants, 184/488 cattle and 59/82 camels. Age distribution in small ruminants ranged from 1 to 11 years, from 1 to 14 years for cattle and from 2 up to 20 years for camels (Supplementary Fig. S2). No correlations of age and seropositivity were observed for small ruminants, cattle and camels, whereas the small number of animals needs to be considered (Fig. 2). In addition, no correlations of ND50 titres with the age of animals (linear regression: small ruminants P = 0·1504, camels P = 0·9979, cattle P = 0·9128) (Supplementary Fig. S3) nor to the regional origin of the animals (logistic regression: small ruminants P = 0·3453, camels P = 0·6502, cattle P = 0·5302) (Supplementary Fig. S4) could be determined.

Fig. 2. Seropositivity of small ruminants, cattle and camels in relation to age of animals. Total numbers of sampled animals per age are depicted by black bars. Red bars indicate numbers of positive animals. The likelihood of seropositivity is not related to age of animals (logistic regression: camels P = 0·917, cattle P = 0·973, small ruminants P = 0·375). For 22/81 samples of camels, 304/488 cattle samples and 143/499 samples of small ruminants the age is not recorded and plotted.
DISCUSSION
In this study we have shown that RVFV-specific antibodies persist in Mauritanian livestock even during inter-epidemic periods and that new infections occur only at a low level by sparse and sporadic transmission to susceptible species. Blood samples from livestock were collected from January to March 2012 and from January to May 2013, thus covering two inter-epidemic periods between three different RVF epidemics. Antibodies were found in all species, yet with significant differences in prevalence (Fisher's exact test, P = 0·03). Small ruminants had the lowest rate (3·8%), followed by cattle (15·4%) and camels (32·0%). These results stand in marked contrast to previous data from Mauritania, obtained from sera collected during acute RVF outbreaks from October to November 2010 [Reference El Mamy14] and later [Reference Jäckel15]. During the RVF outbreak in 2010, antibody prevalence in small ruminants were 43–54% and 33% of tested camels were found to be positive [Reference El Mamy14]. Following the initial outbreak, the study of Jäeckel et al. showed a prevalence of up to 69% of small ruminants, which was markedly higher than those observed for cattle (13%) and camels (45%) [Reference Jäckel15]. Antibody prevalence seems to decrease substantially in small ruminants during inter-epidemic periods, but only marginally in cattle and camels. This may be a result of higher fatality rates in small ruminants during RVFV infections. Moreover, the shorter lifespan of small ruminants causing a faster animal turnover might be a contributing factor. Perhaps the high number of naive sheep and goats contribute to the abortion storms observed even upon re-emerging acute outbreak phases. Similarly low antibody prevalence in small ruminants was also seen in recent studies during inter-epidemic periods in Senegal and Tanzania [Reference Chevalier27–Reference Sumaye29]. Higher prevalence in a given small ruminant population is therefore an indicator for effective epidemic transmission cycles, as the antibody prevalence rises significantly as soon as virus is introduced into the herds. In contrast, higher inter-epidemic prevalence in cattle compared to small ruminants was also found in Senegal and Burkina Faso [Reference Boussini30, Reference Thiongane31]. The general higher prevalence in cattle and camels is probably caused by a combination of lower RVFV case-fatality rates and their longer lifespan. The strong correlation between age and evidence of infection with RVFV was shown in several studies, as older livestock show a significant higher likelihood to be carriers of RVFV-specific antibodies [Reference Sumaye29, Reference Jeanmaire32]. The detected antibodies are presumably caused by previous non-fatal infections. Interpretation of results of camel sera and correlation of seropositivity to age of sampled animals need to be verified by further studies with increased numbers of tested samples.
RVFV-specific antibodies in sheep and goats were mainly detected in Tagant and Gorgol (Table 3). As RVF cases were reported in these areas previously [Reference El Mamy14, Reference Sow16], persisting antibodies are probably causing this observation. The same applies for prevalence detected in camels, as the focus of infection during the outbreak of 2010 was observed in Adrar [Reference El Mamy14]. However, the occurrence of antibodies in cattle was not related to clear regional patterns (P = 0·091). Reasons for that may be the insufficient number of screened cattle sera in the past and the occurrence of subclinical infections, which did not trigger a formal RVF suspicion in cattle. Moreover, movements and trade of animals may have also distorted underlying regional patterns.
In most of the positive samples high neutralizing antibody titres were detected by SNT, which were highest in cattle (ND50 values ranging up to 10 240). A clear regional pattern or correlation with the age of animals was not observed. Differences in ND50 values may be caused by species-specific immunoreactions to RVFV. Additionally, multiple infections may have triggered CD4+ cells and therefore repeatedly elicited neutralizing antibodies [Reference Dodd33, Reference Swain, McKinstry and Strutt34]. Species-specific neutralizing antibody responses against RVFV have not been tested before, but data suggest that there might be a correlation.
SNT and indirect IgG ∆Gn ELISA results demonstrated a high correlation in this study, even with regard to threshold values. However, two sera, which had high percentage of positive control values, contained only a low neutralizing antibody level (ND50 = 10). These results indicate that the generation of antibodies against Gn is not necessarily accompanied by generation of neutralizing antibodies and vice versa. The partially deficient compliance of the indirect IgG ∆Gn ELISA and the commercial cELISA can be explained by utilization of different antigens. As described previously [Reference Swanepoel35], sero-cross-reactivity to other phleboviruses that could cause confusion in the diagnosis of RVFV are not expected.
It has been demonstrated both in infection studies and vaccination trials that IgM antibodies last only for up to 2 months post-infection [Reference Paweska36]. Other reports describe IgM antibodies to persist for up to 5 months post-infection in infected animals [Reference Morvan37]. Notwithstanding, the existence of IgM antibodies is indicative of recent infections. To date, no camelid-specific serological assay for IgM detection is available. Here we report the first application of an indirect IgM in-house ELISA for camelids based on RVFV nucleoprotein and a secondary anti-camelid IgM antibody. Although no IgM-positive camel serum was detected in this study, the indirect IgM ELISA offers a high potential for future monitoring. Screening for the presence of IgM in small ruminants and cattle revealed one positive sample in cattle. The generally low rate of IgM supports the hypothesis that detected antibodies in this study were persisting from previous infections and there was no detectable acute infection circulating in the herds. Comparing the different results of serological tests it is concluded that the IgM-positive cattle sample must have been taken shortly after infection with RVFV, because only IgM and no IgG was detected. The finding was verified by subsequent recovery of partial RVFV genome that can usually be detected for up to 14 days post-infection (dpi) in experimentally infected animals. Concurrent detection of IgM antibodies and viral RNA is possible in a short time-frame of 10 days [Reference Pepin21]. Only limited data from one experimentally infected cattle is available, where viraemia was detectable for 7 dpi and IgM antibodies were detected from 4 dpi onwards [Reference Rippy38]. Hence, it can be assumed that RNA-positive serum of one bovine from Nouakchott indicates the presence of sporadic transmission during enzootic periods in Mauritania. As the sample was taken from a slaughterhouse in Nouakchott, the likelihood that this animal was recently imported from another country is relatively low. In fact it might be explained by commonly existing semi-intensive systems for cattle as described by the FAO [Reference Soule39], mainly characterized by a 1-year period of exploitation of dairy cattle around Nouakchott. To further characterize the potential of those enzootic infections, also in terms of enzootic circulation and maintenance of the virus, prospective studies are needed to analyse both susceptible animals and potential vectors during inter-epidemic periods in detail.
The transition from an endemic pattern to an epidemic outbreak is mainly triggered by additional local environmental and ecological factors that favour mosquito propagation, accompanied by virus replication in the vector and virus transmission. Presumably those supportive factors were absent during this period, thus no transition to severe epidemics occurred.
In conclusion the comparison of the results of our study with data of epidemic periods emphasize the pivotal role of small ruminants as important indicators of acute infections. Since antibody prevalence is characterized as low during inter-epidemic periods, a significant increase could be an early sign of an emerging epidemic.
The results of this study give substantial reasons for small ruminants as sensitive sentinels and main components of an active surveillance program, as neither cattle nor camels are characterized by such remarkable serological differences between epidemic and inter-epidemic period, which is caused by different usage systems and susceptibility to RVFV. Therefore the combined analysis of these different species is suitable to elucidate past and non-recognized RVFV infections of areas without detailed knowledge of RVFV evidence. Data of this study indicate the potential of endemic transmission that should be studied further prospectively. However, for a comprehensive understanding of virus maintenance mechanisms, the analysis of indigenous mosquito populations and corresponding virus dissemination is indispensable. To further elucidate RVFV circulation and transmission in Mauritania, examination and monitoring of mosquitoes and other potentially competent vectors should be included.
SUPPLEMENTARY MATERIAL
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268816003022.
ACKNOWLEDGEMENTS
We thank Birke Böttcher and Tobias Winterfeld for their technical support in all respects. We also thank Dr Andreas Pauly from Tierpark Berlin for providing camel sera.
This work was supported by the German Federal Foreign Office (grant no. 2513AA0374).
DECLARATION OF INTEREST
None.








