Introduction
The cell membrane is an essential component in biology, which plays a vital role in manipulating cellular signaling and communications (Grecco et al., Reference Grecco, Schmick and Bastiaens2011). However, the detailed mechanism of how cells organize these precise membrane interactions and signaling patterns are still largely unknown. It is important to develop advanced tools that can be used to understand the properties and roles of the cell membranes. Meanwhile, molecules and approaches that can regulate membrane functions are also critical tools in medicine and diagnostics.
Different kinds of molecules have been developed to directly modify onto the cell membranes or bind targeted membrane components (e.g. proteins, lipids, and carbohydrates). A variety of small-molecule drugs, aptamers, antibodies, or biorthogonal membrane conjugation approaches have been applied for this purpose (Zhou and Rossi, Reference Zhou and Rossi2014; Xu et al., Reference Xu, Ho, Zhang, Bertrand and Farokhzad2015; Idiago-López et al., Reference Idiago-López, Moreno-Antolín, de la Fuente and Fratila2021). Recently, a type of amphiphilic lipid-DNA conjugates has emerged as an effective and highly flexible tool to modify the membranes of living cells (Bagheri et al., Reference Bagheri, Shafiei, Chedid, Zhao and You2019b; Zhao et al., Reference Zhao, Tian, Bagheri and You2020b). In these conjugates, a lipid moiety is used to anchor into the cell membranes via hydrophobic interactions. Whereas a DNA oligonucleotide is conjugated to provide desired functions on the cell membranes (Schoenit et al., Reference Schoenit, Cavalcanti-Adam and Göpfrich2021). The reason that the DNA strand is a proper choice here is because its structure and interaction can be precisely and programmably controlled via sequence-specific hybridization. In addition, a large variety of functional moieties can be easily conjugated within these DNA oligonucleotides. As a result, these lipid-DNA conjugates have now opened an entirely new field of study, in which we can better understand and exert more control over the structures and functions of the cell membrane.
There are several excellent reviews on the formation of different membrane DNA nanostructures based on these lipid-DNA conjugates, as well as their biomedical and analytical applications (Bagheri et al., Reference Bagheri, Shafiei, Chedid, Zhao and You2019b; Shen et al., Reference Shen, Grome, Yang and Lin2020; Zhao et al., Reference Zhao, Tian, Bagheri and You2020b; He et al., Reference He, Liu, Li and Liu2021; Jia et al., Reference Jia, Zhu, Duan and Wu2021; Lanphere et al., Reference Lanphere, Offenbartl-Stiegert, Dorey and Howorka2021; Shi and Wang, Reference Shi and Wang2021). Here in this work, we will instead mainly discuss some recent advancement, especially during the past three years, in understanding the fundamental biophysical features of these membrane-anchored lipid-DNA conjugates, together with their emerging applications in studying and regulating cell membrane biophysics. We believe such a timely review of the biophysical characteristics of this fast-growing interdisciplinary research field is important for guiding future development of more powerful lipid-DNA tools for broad cell membrane applications.
Biophysical characteristics of the membrane anchoring of lipid-DNA conjugates
Lipid-DNA conjugates can spontaneously insert onto various cell membranes (Huo et al., Reference Huo, Li, Boersma and Herrmann2019; Bagheri et al., Reference Bagheri, Shafiei, Chedid, Zhao and You2019b; Zhao et al., Reference Zhao, Tian, Bagheri and You2020b). Several recent studies have highlighted the importance of lipid moieties on the anchoring efficiency and accessibility of the conjugates. For example, we have provided a quantitative assessment of the cell membrane insertion kinetics of several lipid-DNA conjugates (Bagheri et al., Reference Bagheri, Chedid, Shafiei, Zhao and You2019a). Our results indicated that the cell membrane insertion and departure rates of the conjugates are both strongly regulated by the hydrophobicity of the lipids. To achieve long-term cell membrane modification (>24 h), excess lipid-DNA conjugates and endocytosis inhibitors can be added in the solution. In another study, Howorka and colleagues examined the impact of hydrophobicity and modification position of alkyl chains on the insertion of DNA probes onto both synthetic and living cell membranes (Jones et al., Reference Jones, Joshi, Terry and Howorka2021). Based on experimental data and atomistic molecular dynamics simulation results, they determined that, when these hydrophobic tags are modified at the terminus, the lipid-DNA conjugates are mostly anchored at the outer leaflet of the membranes. Very interestingly, in case of central positioning of the alkyl chains, there is a potential for the DNA strands to span the hydrophobic membranes (Fig. 1a). These membrane-spanning lipid-DNA conjugates can be likely useful for applications in transmembrane deliveries and inner leaflet studies.
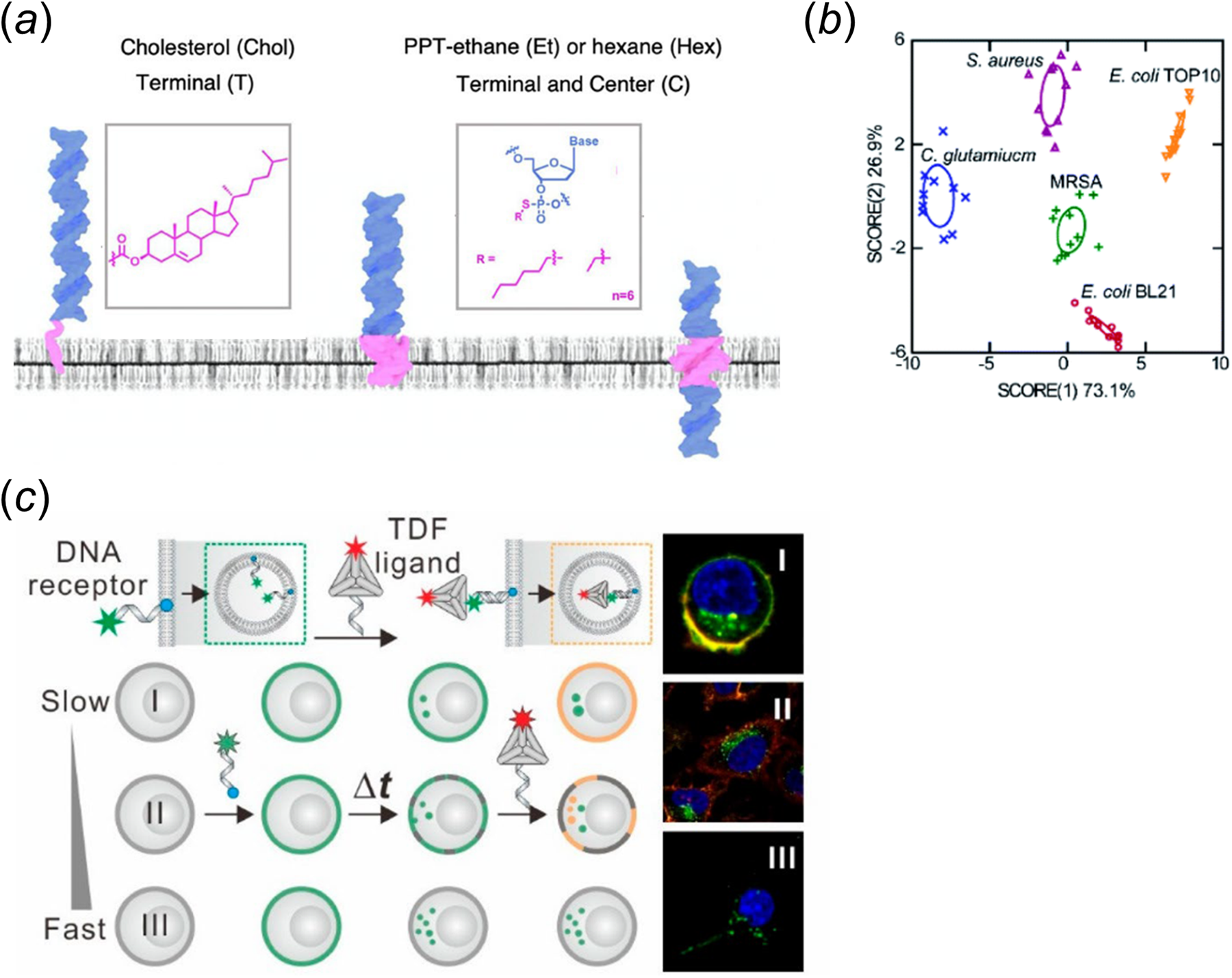
Fig. 1. (a) The impact of lipid type and modification position on the membrane insertion pattern of lipid-DNA conjugates. Adapted with permission from Jones et al. (Reference Jones, Joshi, Terry and Howorka2021). (b) Lipid-DNA conjugates-based linear discriminant analysis on five bacterial strain types (Tian et al., Reference Tian, Bagheri, Keshri and You2021). (c) Identification of cells based on the pattern of probe internalization rate. Reprinted with permission from Zhang et al. (Reference Zhang, Wang, Li and Zhu2020).
Rather than studying the effect of lipid moieties, the Keyser group has assessed the importance of divalent cations, such as Mg2+, in promoting the membrane insertion of lipid-DNA conjugates (Morzy et al., Reference Morzy, Rubio-Sánchez, Joshi, Aksimentiev, Di Michele and Keyser2021). Their experimental and simulation results revealed that these divalent cations can be used to minimize the zeta potential between DNA and the membranes, which are required for membrane modification even in the presence of highly hydrophobic lipid tails. The concentration of divalent cations can also be used to modulate the efficacy of lipid-DNA membrane insertion.
Our previous study has also indicated that the self-aggregation of lipid-DNA conjugates will impact their ability to anchor onto cell membranes (Bagheri et al., Reference Bagheri, Chedid, Shafiei, Zhao and You2019a). The monomeric form of the lipid-DNA conjugates, rather than the aggregated conformation, results in a rapid cell membrane insertion. Keyser and colleagues have carried out a more systematic study to assess the factors that influence the aggregation of cholesterol-DNA conjugates (Ohmann et al., Reference Ohmann, Göpfrich, Joshi and Keyser2019). They observed that by placing a single-stranded DNA overhang (especially a poly-T) near the cholesterol tail, the formation of aggregates can be significantly reduced. Meanwhile, guanine bases should be avoided in the adjacent region of the cholesterol, and DNA strands of minimal secondary structure can also reduce the chance of aggregation. These biophysical characteristics of cholesterol-DNA conjugates, i.e. one of the most used commercially available lipid-DNA conjugates, can function as guidelines for the better engineering of lipid-DNA nanostructures.
Lipid-DNA conjugates have often been viewed as a general anchor for the membrane modification on a large variety of cell types. While this universal capability can be advantageous that allows similar lipid-DNA structures to be directly used in different cellular systems, i.e. without the requirement of specific design for each cell type; however, on the other hand, such low specificity on the type of cell membranes will also hinder their applications for targeting particular cell types in a cell mixture. Recently, this idea of lipid-DNA conjugate being a universal membrane anchor has been challenged. Several biophysical studies indicated that the membrane anchoring of lipid-DNA conjugates may also exhibit some level of selectivity. For example, our results showed that depending on the type of lipid anchor, lipid-DNA conjugates can be used to distinguish different Gram-positive and Gram-negative bacterial strains (Fig. 1b) (Tian et al., Reference Tian, Bagheri, Keshri and You2021). The Howorka group also demonstrated that cholesterol-modified DNA nanobarrels can preferentially anchor onto white blood cells in serum, rather than red blood cells (Arulkumaran et al., Reference Arulkumaran, Lanphere, Gaupp, Burns, Singer and Howorka2021). This membrane selectivity can be likely due to differences in the membrane composition and fluidity among cell types.
In addition to these examples of membrane differentiation based on lipids' natural anchoring selectivity, lipid-DNA conjugates can also be engineered to exhibit specific cell membrane modification. As an example, the Tan group has developed an enzymatic dephosphorylation-based approach for targeted anchoring of phosphorylated lipid-DNA conjugates onto cell membranes that express alkaline phosphatase (Jin et al., Reference Jin, He, Zou and Tan2019). In the presence of these alkaline phosphatases, the removal of the phosphate caps in the lipids dramatically increases the hydrophobicity of lipid-DNA conjugates, which further facilitate their cell membrane insertions. In another similar study, the Di Michele lab designed a target-responsive core-shell DNA nanoparticle system (Walczak et al., Reference Walczak, Brady, Mancini and Di Michele2021). Upon binding with a trigger DNA, its hydrophilic DNA corona was released to allow the unprotected cholesterol-DNA conjugates to anchor onto target cell membranes. There is one more example. On the basis of the observations that different cell types exhibit variable rates of endocytosis, Zhu and colleagues developed a two-step cell membrane differentiation approach by first adding a cholesterol-modified DNA strand and then a complementary strand-anchored DNA tetrahedron (Fig. 1c) (Zhang et al., Reference Zhang, Wang, Li and Zhu2020). The tetrahedral DNA can only bind with outer leaflet-anchored cholesterol-DNA strands, but not the ones internalized. As a result, cells with slow endocytosis rate can be specifically modified with the lipid-DNA tetrahedral structures. Although lipid-DNA conjugates have demonstrated some degrees of anchoring specificity, we must emphasize here that the use of these lipid-DNA structures for targeted membrane modifications is still an unmet need, which is lagging far behind those achieved by antibodies and aptamers.
Lipid-DNA conjugates for studying the biophysical features of cell membranes
While still being a recently emerged field, lipid-DNA conjugates have already been used to study several biophysical characteristics of the cell membranes. Herein, we will mainly discuss some recent advances in this area. For example, the Krishnan lab developed a powerful DNA voltmeter, named as Voltair, for measuring cell membrane potentials (Fig. 2a) (Saminathan et al., Reference Saminathan, Devany, Veetil and Krishnan2021). The Voltair consists of a lipid-modified DNA duplex that is conjugated with a voltage-sensitive dye and a reference fluorophore. Localized changes in the membrane potential can be quantitatively imaged based on a ratiometric fluorescence signal. In addition to the plasma membranes, the Voltair can also be used to detect membrane potentials of specific organelles within living cells.

Fig. 2. (a) A lipid-DNA-based membrane potential sensor containing a voltage-sensitive dye and a reference fluorophore (Saminathan et al., Reference Saminathan, Devany, Veetil and Krishnan2021). (b) Regulating the membrane partitioning of DNA duplex between liquid-ordered and disordered domains using cholesterol- and tocopherol-modified DNA strands. Reprinted with permission from Rubio-Sánchez et al. (Reference Rubio-Sánchez, Barker, Walczak, Cicuta and Michele2021). (c) The use of a DNA Zipper probe for measuring cell membrane lipid–lipid interactions and membrane order (Bagheri et al., Reference Bagheri, Ali, Keshri, Chambers, Gershenson and You2022).
Another major focus of lipid-DNA-based biophysical characterizations of cell membranes has been on studying membrane order and lipid domains. These heterogeneous patterns on the cell membranes are known to facilitate the formation of specific membrane signaling complexes to promote preferential membrane interactions (Sezgin et al., Reference Sezgin, Levental, Mayor and Eggeling2017). However, the direct study of these membrane domains on living cells has been challenging, which is mainly due to their dynamic and small-sized structures. Recent advancement indicated that the lipid-DNA conjugates may provide a potential solution for the live-cell study of these lipid domains. As one of the first examples, we previously developed a DNA walker strategy to convert cell membrane lipid domain-mediated transient interactions into fluorescent readouts (You et al., Reference You, Lyu, Han and Tan2017). In another recent example, Di Michele and colleagues applied cholesterol- and tocopherol-modified DNA probes to regulate cargo transport between liquid-ordered and disordered domains (Fig. 2b) (Rubio-Sánchez et al., Reference Rubio-Sánchez, Barker, Walczak, Cicuta and Michele2021). Here, the choice of lipid anchor can be used to determine the specific domain partitioning of the DNA nanostructures. Based on a modular toehold-mediated strand displacement reaction, membrane lateral redistribution of DNA can be programmably regulated on giant unilamellar vesicles.
We recently engineered a lipid-DNA conjugate-based ‘DNA Zipper’ probe for imaging these short-lived lipid domains on live cell membranes (Fig. 2c) (Bagheri et al., Reference Bagheri, Ali, Keshri, Chambers, Gershenson and You2022). In the DNA Zipper, a pair of short complementary DNA strands was used to extend the membrane duration of cholesterol dimerization interactions. These cholesterol–cholesterol interactions are known to be preferably occurred in membrane liquid-ordered domains. These DNA Zipper probes have allowed us to study lipid domain-dependent activation of the T-cell receptor (TCR) signaling. Upon an anti-CD3/anti-CD28 antibody co-stimulation, our results indicated that cell membrane CD3 relocates into the ordered domains, and interestingly, the formation of membrane order and TCR activation happens almost at the same time. It is worth mentioning that this type of DNA Zipper probes may also be potentially applied to measure transient protein–protein or lipid–protein interactions on the cell membranes.
In another attempt to apply the lipid-DNA conjugate for studying lipid domains, Sun et al. used cholesterol-modified DNA duplexes to stabilize lipid domains on the cell membranes (Sun et al., Reference Sun, Su, Wang, Xia, Xu and Li2020b). The authors demonstrated that such stabilization of lipid domains can be used to induce membrane aggregation of the TCR-CD3 complex and to activate T-cell proliferation. These lipid-DNA conjugates can thus be potentially used as an antibody-like stimulator to activate the TCR signaling. In another study, using peptide-major histocompatibility complex- and anti-CD28 antibody-modified cholesterol-DNA conjugates, Sun et al. have further studied the effect of ligand clustering and their vertical distributions on regulating TCR activations (Sun et al., Reference Sun, Shen, Xu, Han, Fan and Liu2020a). In their approach, based on a multivalent biotin–avidin interaction, biotin-labeled cholesterol-DNA conjugates were applied to control the formation of membrane clusters and thus a laterally heterogeneous pattern of the ligands. The authors demonstrated that by mimicking lipid domains to precisely position the membrane ligands, these lipid-DNA conjugates can achieve optimized ability of TCR activation both in vitro and in vivo.
So far, these lipid-DNA-based membrane domain studies have not yet reached single-molecule levels and have not been combined with super-resolution imaging techniques. This can be an important direction that potentially enables the real-time tracking of membrane lipid interactions and for studying the sizes of individual lipid domains and for monitoring their dynamic membrane changes. Meanwhile, studying the membrane preference of lipid–protein interactions can be another interesting direction for applying these lipid-DNA conjugates. These membrane lipid–protein interactions have been until now mostly explored using computational or destructive approaches (Bagheri et al., Reference Bagheri, Ali and You2020). In addition, other than membrane lateral diffusion or interactions, interleaflet coupling is another major process for maintaining the integrity and signaling of the cell membranes (Fujimoto and Parmryd, Reference Fujimoto and Parmryd2017; Doktorova et al., Reference Doktorova, Symons and Levental2020). Lipid-DNA conjugates may potentially also be useful in studying these dysregulated interleaflet coupling processes in diseases such as cancer.
Lipid-DNA conjugates for detecting and regulating intercellular biophysical interactions
Another major biophysical application of the lipid-DNA conjugates is for the detection of intercellular forces. Our group previously developed the first lipid-DNA-based probes for imaging molecular tensions at cell–cell junctions (Zhao et al., Reference Zhao, O'Brien, Mudiyanselage and You2017). Our probe consisted of a ligand-modified short DNA hairpin structure that can be unwound in the event of a strong pulling by receptors on the neighboring cells (Fig. 3a). The opening of the hairpin can further result in the separation between a fluorophore and a quencher, and thus intercellular forces can be converted into fluorescence signals for imaging under fluorescence microscope. Wang et al. have recently used these probes to map the localized intercellular forces during collective cell migration (Wang et al., Reference Wang, Yang, Pan, Kang, Xu and Chen2020). Their results indicated a strong correlation between E-cadherin-mediated intercellular tensile forces and energetic costs of cell migration, especially at the edge of epithelial wound.

Fig. 3. (a) Lipid-modified membrane DNA tension probe for imaging molecular forces at cell–cell junctions. Adapted with permission from Zhao et al. (Reference Zhao, O'Brien, Mudiyanselage and You2017). (b) The design of a ratiometric DNA force probe that is capable of quantifying a large range of intercellular forces (Zhao et al., Reference Zhao, Li, Xie and You2020a).
To further enable the quantitative measurement of intercellular forces, we have lately engineered two ‘next-generation’ lipid-DNA-based mechanical probes (Zhao et al., Reference Zhao, Li, Xie and You2020a; Keshri et al., Reference Keshri, Zhao, Xie and You2021). In our first system, a reference fluorophore was added to normalize the cell membrane distribution variations of the DNA probes (Fig. 3b) (Zhao et al., Reference Zhao, Li, Xie and You2020a). Meanwhile, by incorporating two DNA hairpins together in one construct, a large range of molecular tensions was able to be studied. Based on a ratiometric fluorescence measurement, the cellular distributions of E-cadherin-mediated forces during epithelial adhesion and migration were quantitatively determined. In our second approach, quantification of intercellular forces was achieved through measuring fluorescence lifetime rather than fluorescence intensity (Keshri et al., Reference Keshri, Zhao, Xie and You2021). These fluorescence lifetime signals are independent on the probe concentration, and as a result, force measurement can be achieved without the requirement of a reference fluorophore. A simultaneous detection of multiple mechanotransduction ligand–receptor pairs can be much more easily achieved. For example, our results indicated that during an epithelial-to-mesenchymal transition process, a gradually increased contribution of N-cadherin tensions and a correspondingly decreased E-cadherin force occurs.
There remain a lot of potential avenues for progress in this emerging field. For example, current lipid-DNA conjugates have mostly been used in a 2D cell culture system, whereas the physiological pattern of the forces can be quite different from that in a real 3D network. Furthermore, these lipid-DNA conjugates can be quickly internalized or dissociated from the cell membranes, which is why these force measurements are often conducted within a relatively short time window. It would be a great benefit to develop lipid-DNA probes of prolonged membrane stabilities to allow for long-term monitoring the dynamics of intercellular forces.
In addition to these force measurement, lipid-DNA conjugates have also been extensively applied to bring cells into physical proximity for regulating cellular aggregations. For example, in one early study, based on DNA hybridization, the Hook group used cholesterol-DNA probes to fuse synthetic lipid vesicles together (Stengel et al., Reference Stengel, Zahn and Höök2007). The similar approach was later used to promote small molecule deliveries between synthetic vesicles (Simonsson et al., Reference Simonsson, Kurczy, Trouillon, Hook and Cans2012). Moreover, lipid-DNA conjugates enabled the modification of aptamers onto live cell membranes, which further advanced our ability to control specific cellular aggregation and communication (Xiong et al., Reference Xiong, Liu, Zhao and Tan2013). These early demonstrations laid the foundation for the following recent progress in modulating intercellular communications through versatile design of DNA nanostructures.
For example, the Tan group recently demonstrated a membrane-anchored multivalent DNA nanostructure to promote the efficiency of intercellular communication and aggregation (Li et al., Reference Li, Xun, Zheng, Peng, Qiu and Tan2021). This nanostructure was synthesized based on a tandem hybridization chain reaction (Fig. 4a) and can be modularly designed to respond to molecular triggers (e.g. ATP) and induce a targeted cell therapy. They have also similarly used such hybridization chain reaction and a cholesterol-modified DNA triangular prism to synthesize multi-layered DNA network on the cell surfaces (Fig. 4b) (Guo et al., Reference Guo, Zhang, Yang and Tan2022). As a result of this large DNA network, controllable cell aggregation and intercellular molecular transportation was able to be achieved. Membrane-anchored hybridization chain reaction has also been used in another study by Wang and colleagues to improve the proximity between natural killer cells and cancer cells (Shi et al., Reference Shi, Wang, Davis and Wang2020). An aggregation-enhanced killing of target cancer cells was reported. As shown by these examples, lipid-DNA-mediated specific and controllable contact of cells can be potentially served as a new therapeutic strategy.

Fig. 4. (a) Hairpin chain reaction-based programming of intercellular interactions and aggregations. Reprinted with permission from Li et al. (Reference Li, Xun, Zheng, Peng, Qiu and Tan2021). (b) A cell surface DNA scaffold with tunable thickness (Guo et al., Reference Guo, Zhang, Yang and Tan2022). (c) Programming-specific cell–cell interactions based on a DNA strand displacement reaction. Reprinted with permission from Xiao et al. (Reference Xiao, Lai, Yu and Pei2021). (d) Studying distance-dependent fusion and lipid exchange between two DNA-tethered liposomes (Bian et al., Reference Bian, Zhang, Xiong, De Camilli and Lin2019).
Most of the above-mentioned examples used cholesterol-modified DNAs to regulate intercellular interactions, in another report, Ren and colleagues have instead demonstrated that glycolipid- and glycoprotein-modified DNA nanostructures can also be used for cell membrane modification and regulating multicellular aggregation (Wijesekara et al., Reference Wijesekara, Liu, Wang and Ren2021). These glycol-conjugated DNA strands exhibit tunable accessibility and distance from the plasma membrane, which can be useful for controlling the efficiency of cell assemblies.
Lipid-DNA-based logic control over these cell aggregations have also been separately demonstrated by the Lu group and Pei group (Qian et al., Reference Qian, Zhou, Guo, Wu, Yang and Lu2021; Xiao et al., Reference Xiao, Lai, Yu and Pei2021). In Pei's approach, a structural switchable DNA probe was modified on the membranes of natural killer cells. Depending on the sequence of the input trigger DNA, this DNA probe can form different three-way junction structures to induce the selective aggregation of natural killer cells with either Jurkat or K562 cells (Fig. 4c) (Xiao et al., Reference Xiao, Lai, Yu and Pei2021). While in Lu's strategy, two metal ions (e.g. Mg2+ and Zn2+) were used to trigger the activation of DNAzymes and thus achieve controllable cellular disaggregation (Qian et al., Reference Qian, Zhou, Guo, Wu, Yang and Lu2021). These logically controlled systems demonstrated that lipid-DNA conjugates can potentially function as a versatile and smart tool to regulate intercellular network.
Environmental pH has also been applied to regulate lipid-DNA-mediated cell aggregations (Hou et al., Reference Hou, Zhu, Zhao and Li2021). Normally, a pH-dependent switch between DNA duplex and an i-motif or triplex structure was used to realize such pH control. These pH-responsive DNA nanodevices have also been applied to regulate the anchoring of cell-derived nanovesicles onto target cell membranes, which can be potentially useful as an alternative therapeutic delivery strategy (Huang et al., Reference Huang, Guo, Zhang and Tan2021a).
In addition to regulating living cell aggregations, lipid-DNA conjugates have also been used to programmably control the interactions and communications among artificial cells (Bian et al., Reference Bian, Zhang, Xiong, De Camilli and Lin2019; Qiu et al., Reference Qiu, Li, Du and Choi2021). In one interesting example, the Lin group demonstrated that DNA origami can be used to precisely modulate the distance between two liposomes, and as a result, they were able to study distance-dependent dynamics of fusion and lipid exchange between two membrane bilayers (Fig. 4d) (Bian et al., Reference Bian, Zhang, Xiong, De Camilli and Lin2019). Meanwhile, these membrane-anchored DNA origami have also been used to separate liposomes into highly homogenous fractions with specific sizes (Yang et al., Reference Yang, Wu, Wang and Lin2021b).
There are several advantages in using lipid-DNA conjugates for the programming and modulation of cell–cell interactions. First, this type of membrane modification and cell aggregation process is generally fast, and the membrane density and thickness of lipid-DNA conjugates can be easily controlled. As a result, it is possible to regulate the degree and distance of intercellular interactions. In addition, lipid-DNA can be a useful system for understanding intercellular communications and signaling, and also potentially applied for cell therapy. However, several factors need to be considered before possibly applying this DNA network to generate real 3D tissues or complex cell structures. For example, the lipid-DNA conjugates can dissociate from the cell membranes or get degraded and thereby the cell aggregation may be lost over time. Meanwhile, it is still challenging to aggregate specific types of cells in a mixture due to limited specificity and persistence of lipid-DNA-based membrane modification. More fundamental studies on the biophysical features of lipid-DNA conjugates are still required to allow the practical usage of these programmable, modular, and easy-to-use tools for regulating intercellular interactions in tissue engineering and targeted therapy.
Lipid-DNA conjugates for detecting localized membrane signaling
Lipid-DNA conjugates have also found several unique applications in detecting and imaging signaling molecules near the cell membranes. These membrane-localized measurements are important for studying the sources, distributions, and kinetics of signaling generation and transduction. As one of the first examples, the Tan group demonstrated the use of lipid-modified DNAzyme to detect membrane-localized metal ions (Qiu et al., Reference Qiu, Zhang, Jiang and Tan2014). Similarly, other DNA structures such as G-quadruplex can also be used in the detection of membrane potassium ions (Xiong et al., Reference Xiong, Zhu, Rong and Tan2016). In more recent years, several interesting stimuli-responsive DNA probes have been further engineered for the membrane detection of various target analytes. For example, Liu et al. developed a membrane pH sensor based on cholesterol-modified DNA tetrahedral structure (Fig. 5a) (Liu et al., Reference Liu, Li, Li and Ruan2021). This membrane-anchored DNA probe contained a pH-sensitive i-motif and can be used to quantitatively image a large range of pH changes near the cell membranes. In another example, Zeng et al. anchored an aptamer-modified cholesterol-DNA triangular prism onto neuroblastoma cells and applied it to detect the cellular release of dopamine, a neurotransmitter (Zeng et al., Reference Zeng, Wang, Xie and Wang2020). The Krishnan lab has recently developed a pair of quantitative DNA-based nitric oxide sensors for detecting the activity of nitric oxide synthase 3 at both plasma membrane and trans-Golgi network (Fig. 5b) (Jani et al., Reference Jani, Zou, Veetil and Krishnan2020). These localized sensors are indeed powerful tools for spatially mapping and comparing enzymatic activities at specific locations in living cells.

Fig. 5. (a) An DNA i-motif-based sensor for sensing a broad range of pH changes on cell membranes. Reprinted with permission from Liu et al. (Reference Liu, Li, Li and Ruan2021). (b) DNA-based nitric oxide sensors for detecting NOS3 activities at both plasma membrane and trans-Golgi network (Jani et al., Reference Jani, Zou, Veetil and Krishnan2020). (c) Membrane-anchored DNA probe for monitoring the cellular release of ATP during apoptosis (Zheng et al., Reference Zheng, Wang, Shi, Peng, Shi and Li2021). (d) Design of fusogenic vesicles that can induce lipid-DNA modification on both inner and outer leaflets of the cell membranes (Lin et al., Reference Lin, Chen, Zhao, Tang, Nie and Xing2022).
In another interesting study, Li and colleagues developed a membrane-anchored logic-gated DNA nanodevice that can be used to monitor the cellular release of ATP under specific pH and potassium ion conditions (Zheng et al., Reference Zheng, Wang, Shi, Peng, Shi and Li2021). This nanodevice combined a pH-sensitive DNA triplex, a K+-responsive G-quadruplex, and an ATP-binding aptamer (Fig. 5c). Only in the presence of both H+ and K+ ions, the DNA device can be activated for the sensing of ATP on target cell membranes. Instead of using aptamers, the Li group demonstrated that the cell membrane detection of ATP can also be achieved based on DNAzymes (Zhao et al., Reference Zhao, Chang, Zhang and Li2021). Upon binding with ATP, the DNAzyme can catalyze a nucleophile attacking of its own 5’-hydroxyl group on the ATP phosphate, which results in a self-phosphorylation reaction and a λ-exonuclease-based cleavage of the DNAzyme strand to generate an activated fluorescence signal.
The detection of ATP has also been achieved from the inner leaflet of the cell membranes. Xing and colleagues recently developed a really clever strategy to modify lipid-DNA conjugates onto both inner and outer leaflets of the cell membranes (Fig. 5d) (Lin et al., Reference Lin, Chen, Zhao, Tang, Nie and Xing2022). In this system, by incorporating lipid-DNA conjugates in both inner and outer surface of a liposome, upon the fusion of the liposome with the cell membrane, lipid-DNA can modify onto both leaflets. This is an important advancement since most previous lipid-DNA conjugates can only be applied on the outer leaflet of the plasma membrane. Now membrane localized signaling, from both sides of the surface, can be potentially studied with these lipid-DNA conjugates.
In addition to just visualizing localized membrane signaling, the Yang lab developed a cholesterol-DNA conjugate that is capable of both imaging and modulating cell signaling processes (Chen et al., Reference Chen, Xu, Li and Yang2022). This system consisted of two membrane-anchored aptamer arms: a regulating arm to inhibit c-MET dimerization and an imaging arm to detect the released VEGF after the activation of c-MET dimerization. The regulating arm can further be controlled by light for designing a functional multitasking membrane-anchored DNA nanodevice.
Considering the existence of many target-binding aptamers and other functional DNA moieties, we expect more diverse types of membrane signaling analytes can be potentially detected using these modular lipid-DNA conjugates. Before these signaling molecules diffuse and get diluted in the bulk media, the relatively high target concentration around the cell surfaces likely enables these membrane-anchored lipid-DNA probes to be highly sensitive. In addition to the plasma membranes, the ability of these lipid-DNA conjugates to anchor onto membranes of different intracellular organelles has further extended their applications in studying membrane signaling (Saminathan et al., Reference Saminathan, Devany, Veetil and Krishnan2021). Meanwhile, we expect more advanced lipid-DNA structures will continuously emerge with minimized internalization and improved membrane stability for long-term membrane detections.
Lipid-DNA conjugate for regulating membrane transport
Another important feature of the plasma membrane is to function as a boundary for controlled and selective molecular transportation. For example, lipid-anchored DNA helix bundles have been known to form nanochannels and span synthetic membranes (Langecker et al., Reference Langecker, Arnaut, Martin and Simmel2012; Burns et al., Reference Burns, Seifert, Fertig and Howorka2016). These membrane-anchored lipid-DNA conjugates have been further used as artificial transmembrane transporters (Thomsen et al., Reference Thomsen, Malle, Okholm and Kjems2019; Lanphere et al., Reference Lanphere, Offenbartl-Stiegert, Dorey and Howorka2021). For example, Dash and colleagues developed an artificial ion transport channel composed of lipid-modified guanosines and a human telomeric G-quadruplex DNA strand (Debnath et al., Reference Debnath, Chakraborty, Kumar, Chaudhuri, Jana and Dash2020). This membrane-anchored DNA complex was capable of selectively transporting K+ ions across both artificial membranes and live mammalian cell membranes. Meanwhile, this artificial membrane channel can be easily switched on and off over a few cycles by adding high-affinity ligands such as cytosine or thiazole orange to displace the lipid-modified guanosines.
These lipid-DNA-based membrane channels have also been used to design artificial cells. In one example, the Tan group has used a cholesterol-modified six-helix DNA bundle to achieve reaction regulation in a biomimetic giant vesicle (Peng et al., Reference Peng, Xu, Wang and Tan2020). In the presence of ATP, this barrel-like membrane channel can be selectively activated by releasing a DNA locker strand made of an ATP-binding aptamer (Fig. 6a). As a result, metal ions, e.g. Zn2+, can be transported through the membrane and trigger an artificial DNA feedback loop inside the vesicle. Interestingly, this feedback loop can further be engineered to generate a blocker DNA strand to switch the membrane DNA channel back to the off state. To mimic intercellular signal transduction, the Tan group has also engineered another synthetic transmembrane channel on giant membrane vesicles. This membrane channel can be activated by another DNA nanostructure on neighboring vesicles (Fig. 6b) (Yang et al., Reference Yang, Guo, Liu and Tan2021a). In this system, interactions among vesicles can trigger the release of a blocker DNA via strand displacement reactions, resulting in the activation of membrane channel for the ion transportation.
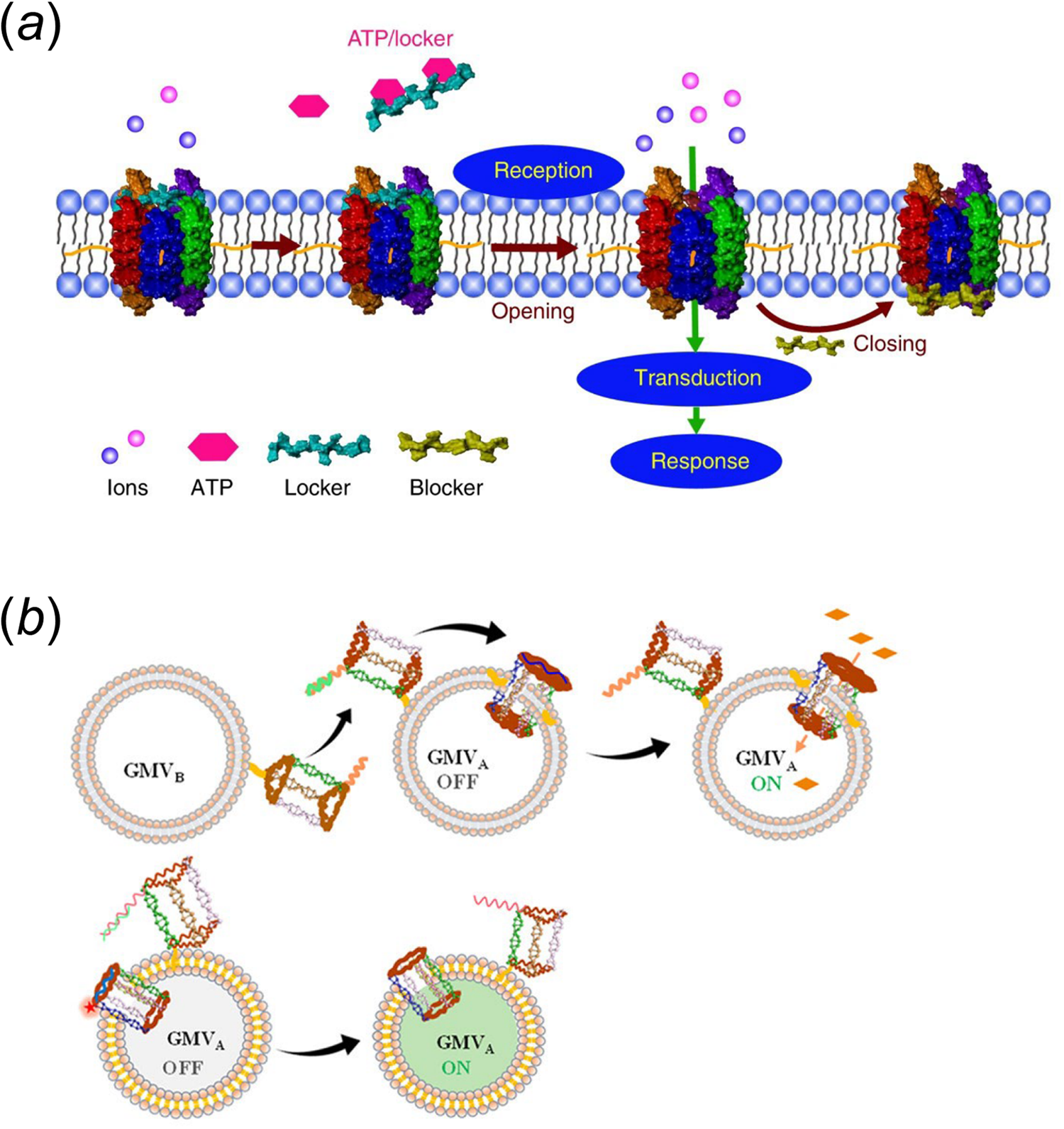
Fig. 6. (a) An ATP stimuli-responsive barrel-like membrane DNA channel that is capable of inducing downstream signaling cascades (Peng et al., Reference Peng, Xu, Wang and Tan2020). (b) A lipid-DNA nanoprism that can insert into membranes as a nanopore when interacting with nearby vesicles. Reprinted with permission from Yang et al. (Reference Yang, Guo, Liu and Tan2021a).
Another important potential application of these artificial membrane channels is for therapeutics. Cell membrane-anchored DNA origami-based nanopores have been shown to be able to enhance the membrane permeability and lead to apoptosis and cytotoxicity (Burns et al., Reference Burns, Al-Juffali, Janes and Howorka2014; Kocabey et al., Reference Kocabey, Kocabey, Schneiter and Rüegg2021). Activatable DNA-based nanochannels have also been used for targeted cancer therapy. For example, Chen et al. have designed a DNA nanopore-anchored fusogenic liposome that can only be activated in tumor-like acidic conditions and then fuse with the cell membranes to induce pyroptosis-like cell death (Chen et al., Reference Chen, Liang, Chen, Wu and Zhang2020).
While the ideas of using artificial DNA-based membrane channels to regulate transmembrane transport have been demonstrated on both synthetic vesicles and real cells, the ability of tuning and modulating the behaviors of these membrane nanopores to enable real biomedical usage is still under exploration. The current generation of DNA membrane channels has limited specificity regarding what target molecules can be transported and what types of cells are modified. Nevertheless, this is a very promising future direction to really demonstrate the therapeutic power of these membrane-anchored DNA nanostructures.
It is worth mentioning that in addition to these artificial membrane channels, some recent studies also showed that these lipid-DNA conjugates can be directly used for transmembrane transport in vivo. Two very exciting reports by the Yokota group found that cholesterol- and tocopherol-modified DNA/RNA heteroduplexes are highly promising tools to effectively deliver antisense oligonucleotides and knock down genes inside rodent central nervous system and lymphocytes (Nagata et al., Reference Nagata, Dwyer, Yoshida-Tanaka and Yokota2021; Ohyagi et al., Reference Ohyagi, Nagata, Ihara and Yokota2021). While the detailed mechanism of such high-efficacy transmembrane delivery still requires further studies.
Conclusion
In this review, we have highlighted some recent advancement in studying the biophysical features and membrane applications of the lipid-DNA conjugates. These conjugates are relatively easy to synthesize and can be modified with a variety of functional units. Meanwhile, the programmable nature of DNA sequences has provided additional functionalities and benefits that allow the versatile usage of these conjugates, without the requirement of genetic encoding. Recently, there are several major breakthroughs in this field of research. These lipid-DNA conjugates have been used to measure and regulate various biophysical features of the cell membranes, including mechanical forces, voltages, lipid domains, and transport. The recent ability to modify these conjugates on the inner leaflets of cell membranes also provides a new avenue for studying inner membrane biophysical phenomena and for controlling membrane signaling network. Given the current interest in artificial and synthetic cellular systems, lipid-DNA conjugates have also provided an effective strategy in biomimicry and synthetic biology.
In addition to the examples discussed above, we think there are several other prospective areas that these lipid-DNA conjugates may make a significant contribution. For example, these conjugates can be used as a barcode to label specific types of cells in a mixture, which may be used for cell sorting or locating in single-cell omics studies (McGinnis et al., Reference McGinnis, Patterson, Winkler and Gartner2019; Huang et al., Reference Huang, Chen, Shen and Li2021b). In addition, these lipid-DNA structures may play a more important role in cell engineering for cancer immunotherapy. Furthermore, the use of lipid-DNA conjugates for the LEGO-like bottom-up assembly of tissues and organs remains an ambitious but exciting direction.
While general limitations do exist in this emerging field, such as the rapid internalization, limited membrane specificity, and inability for long-term membrane applications, recent progress made in the field has been tremendous. Innovative strategies to further address these challenges will be highly impactful. With insights into the current progress and challenges, we hope this review will motivate future advancement of lipid-DNA conjugates toward more exciting cellular applications and for solving fundamental membrane biophysical questions.
Acknowledgements
The authors gratefully acknowledge NIH R35GM133507, Camille Dreyfus teacher-scholar award, and a start-up grant from UMass Amherst to M. You. We also thank every other member of the You Lab for useful discussion.
Conflict of interest
None.