To the Editor—Pseudomonas aeruginosa is an opportunistic pathogen involved in a wide variety of infections among hospitalized patients; it is one of the main agents that cause pneumonia in mechanically ventilated patients.Reference Aaron, Kottachchi and Ferris1 After colonizing the respiratory tract, P. aeruginosa may lead to extensive damage to the host tissues via the production of virulence factors, which are controlled by the quorum-sensing (QS) mechanism, an important cell-to-cell communication system for biofilm formation and maintenance.
As previously reported,Reference Latifi, Foglino, Tanaka, Williams and Lazdunski2, Reference Perez, Machado and Barth3 at least 2 major pathways, the las and rhl systems, are involved in a biofilm regulation process through the production of signaling molecules (ie, acyl-homoserine lactones; AHLs). In many cases, P. aeruginosa is an antibiotic-sensitive strain, but clinical concern arises when an infection involving biofilm needs to be treated. In addition, widely used anti-Pseudomonas antibiotics, such as meropenem, can be affected from the standpoint of resistance development when a biofilm is involved. By comparing the minimal inhibitory concentration (MIC) and minimal biofilm eradication concentration (MBEC), we aimed to evaluate (1) the influence of presence of the las and rhl genes on ability to produce biofilm and (2) the increase in meropenem resistance.
In total, 199 P. aeruginosa isolates recovered from endotracheal secretions of hospitalized patients, collected between January 2015 and July 2016, were included in this study. The isolates were identified based on their inability to ferment glucose, on their ability to production a blue–green pigment, and on biochemical tests such as oxidase production, nitrate reduction, and growth on cetrimide agar (bioMérieux, Marcy l’Etoile, France). As inclusion criteria, we selected only isolates (1 per patient) shown to be meropenem susceptible because we aimed to compare the impact of biofilm production on the development of resistance to meropenem.
Biofilm production was performed according to a microtiter plate assay, and polymerase chain reaction (PCR) assays were carried out to determine the presence of lasI, lasR, rhlI, and rhlR genes using specific primers, according to parameters previously described.Reference Perez, Machado and Barth3 The minimal inhibitory concentration (MIC) was determined using microtiter plate assays as recommended by the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI 2018). The minimal biofilm eradication concentration (MBEC) assay was performed as described by Moskowitz et al Reference Moskowitz, Foster, Emerson and Burns4
The presence of genes related to the QS system was evaluated by PCR for 199 P. aeruginosa isolates. In 161 of these 199 isolates (80.9%), all 4 QS genes evaluated were detected (Table 1). In 8 of the non–biofilm-producing isolates, no QS genes were detected. In 3 other isolates, only genes from the rhL system were detected. Thus, we hypothesized a central role of the las system, mainly, on the biofilm development and/or structure maintenance. Regarding biofilm producers, 188 of 199 isolates (94.5%) produced any degree of biofilm (Table 1). Importantly, most isolates were moderate or strong biofilm producers, and this finding probably reflects a high meropenem resistance level because high values of MBEC (ie, high levels of resistance) were observed among these isolates (Table 1). In fact, high MBEC/MIC ratios of 1,000× (0.064/64) were found in 12 strong-biofilm–producing isolates. The differences between the values of the MBEC and MIC were more evident for isolates with a moderate or strong ability to produce biofilms, whereas non–biofilm-producing isolates showed the same MBEC and MIC values.
Table 1. Quorum-Sensing (QS) Genes, Ability to Produce Biofilm and Minimal Inhibitory Concentration (MIC) and Minimal Biofilm Eradication Concentration (MBEC) Among 199 Meropenem-Susceptible P. aeruginosa Isolates
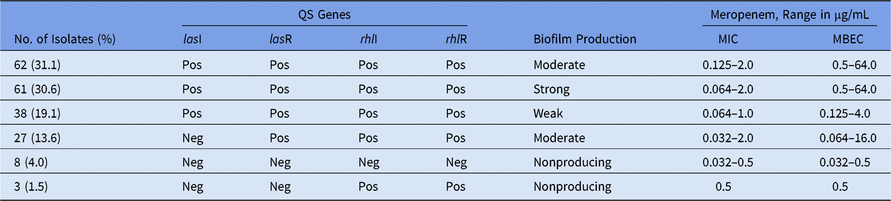
Note. Pos, positive; Neg, negative.
Biofilm formation was largely associated with infections due to colonization of medical devices (eg, catheters and tracheostomy tubes). In these cases, eradication of the infection is difficult because the antimicrobial agents may not penetrate the biofilm, and the decreased metabolic activity of bacteria within biofilms is also due to the increase in gene transfer.Reference Stewart5 In a prior study, we demonstrated that P. aeruginosa isolates harboring metallo-β-lactamases had the ability (most strong or moderate) to produce biofilm in vitro, which represents an “overlapping of mechanisms” that challenges pulmonary infection treatment.Reference Perez, Antunes, Freitas and Barth6 On the other hand, in another study, Acinetobacter baumannii complex showed an inverse relationship between meropenem resistance and biofilm formation.Reference Perez7 Thus, it is important to evaluate the behavioral relation of each bacterial species to better establish targeted prevention efforts and control measures. Although attention has been focused on the widespread resistance to carbapenems, little is known about biofilm production and its important repercussions. Biofilm may represent an important bacterial barrier for control and prevention measures, even for susceptible strains.
To the best of our knowledge, this is the first study evaluating the presence of QS genes among P. aeruginosa clinical isolates and its influence on biofilm development and meropenem resistance. Our results have demonstrated that isolates planktonically susceptible to meropenem (under laboratory conditions) may show high levels of resistance to this drug if they occur in biofilm. This finding may reflect a nonresponse to infection control measures based only on standard susceptibility profiles, and greater caution should be taken when biofilm-related infections are suspected.
Acknowledgments
The author would like to thank Sophia L. Perez for technical support.
Financial support
No financial support was provided relevant to this article.
Conflicts of interest
All authors report no conflicts of interest relevant to this article.



