Herbicide resistance continues to pose a significant threat to the sustainability of cropping systems worldwide. In particular, the evolution of resistance to the active ingredient glyphosate has reduced the effectiveness of glyphosate-tolerant systems. Since the first report of glyphosate resistance in a Conyza canadensis (L.) Cronq. accession from Delaware (VanGessel Reference VanGessel2001), there have been an additional 16 weed species with confirmed resistance to glyphosate spanning 38 states in the United States (Heap Reference Heap2017). In Canada, the initial discovery of glyphosate resistance was reported for giant ragweed (Ambrosia trifida L.) in 2008 (Vink et al. Reference Vink, Soltani, Robinson, Tardif, Lawton and Sikkema2012). Subsequently, four additional weed species, including C. canadensis have developed glyphosate resistance in Canada, three of which originate from the most southwesterly portion of the province of Ontario bordering the United States (Heap Reference Heap2017).
In order to exert its lethal action on plants, glyphosate must first be absorbed through the leaf cuticle and reach the parenchymal cells (Caseley and Coupland Reference Caseley and Coupland1985; Kirkwood et al. Reference Kirkwood, Hetherington, Reynolds and Marshall2000). Following loading in the phloem, it is translocated to various sink tissues, where it accumulates (Denis and Delrot Reference Denis and Delrot1993; Gougler and Geiger Reference Gougler and Geiger1981). Inhibition of the target site, the chloroplastic, nuclear-encoded enzyme 5-enolypyruvylshikimate-3-phosphate synthase (EPSPS), occurs after enough herbicide has entered the chloroplasts (Steinrucken and Amrhein Reference Steinrucken and Amrhein1980). Following inhibition, the concentration of arogenate, an intermediate in the synthesis of aromatic amino acids, decreases, causing a loss of regulatory feedback inhibition to DAHP, an enzyme preceding EPSPS. This results in shortages of carbon for other essential pathways (Siehl Reference Siehl1997) and is thought to be the main cause of glyphosate’s phytotoxic action.
The molecular mechanism(s) conferring glyphosate resistance in C. canadensis have received a great deal of attention (Dinelli et al. Reference Dinelli, Marotti, Bonetti, Minelli, Catizone and Barnes2006; Feng et al. Reference Feng, Tran, Chiu, Sammons, Heck and CaJacob2004; Ge et al. Reference Ge, d’Avignon, Ackerman and Sammons2010; González-Torralva et al. Reference González-Torralva, Rojano-Delgado, de Castro, Mülleder and De Prado2012; Peng et al. Reference Peng, Abercrombie, Yuan, Riggins, Sammons, Tranel and Stewart2010, Reference Peng, Lai, Lane, Nageswara-Rao, Okada, Jasieniuk, O’Geen, Kim, Sammons, Rieseberg and Stewart2014; Yuan et al. Reference Yuan, Abercrombie, Cao, Halfhill, Zhou, Peng, Hu, Rao, Heck, Larosa, Sammons, Wang, Ranjan, Johnson, Wadl, Scheffler, Rinehart, Trigiano and Stewart2010). Since the first report of glyphosate-resistant (GR) C. canadensis in 2001, accessions from across the United States have been extensively studied and, thus far, all have demonstrated non–target site mediated resistance to glyphosate (Dinelli et al. Reference Dinelli, Marotti, Bonetti, Minelli, Catizone and Barnes2006; Ge et al. Reference Ge, d’Avignon, Ackerman and Sammons2010; Moretti and Hanson Reference Moretti and Hanson2017). In 2004, Feng et al. reported that glyphosate resistance in C. canadensis was associated with reduced translocation of glyphosate to the target site. A subsequent study by Ge et al. (Reference Ge, d’Avignon, Ackerman and Sammons2010) identified the selective sequestration of glyphosate into the vacuole as the molecular mechanism reducing translocation and conferring glyphosate resistance in that species. Aside from Dinelli et al. (Reference Dinelli, Marotti, Bonetti, Minelli, Catizone and Barnes2006), who observed a small increase in the expression of EPSPS in addition to reduced translocation in GR C. canadensis accessions, no other mechanisms conferring glyphosate resistance have been reported in this species to date.
Concerns over the geographic spread of GR C. canadensis have centered on the unique propensity of this species for long-distance seed dispersal. Conyza canadensis is a primarily self-pollinating species that can produce up to 200,000 seeds plant−1 (Weaver Reference Weaver2001). The dispersal of these propagules is wind assisted, with a potential range of 500 km from the source accession (Shields et al. Reference Shields, Dauer, VanGessel and Neumann2006). While the vast majority of C. canadensis seed disperse no farther than 100 m from the parent plant (Dauer et al. Reference Dauer, Mortensen and VanGessel2007), the incremental spread of a GR biotype across county or state lines is a realistic possibility. In this respect, the evolution of glyphosate resistance in C. canadensis has provided an interesting case study on the balance between the independent selection for herbicide resistance within and among regions versus the long-distance dispersal of resistant propagules from neighboring accessions. At the state level, Dinelli et al. (Reference Dinelli, Marotti, Bonetti, Minelli, Catizone and Barnes2006) concluded that, while GR C. canadensis accessions from Delaware, Virginia, Ohio, and Arkansas shared common non–target site resistance mechanisms, they did not share a common evolutionary or geographic origin. Similarly, Okada et al. (Reference Okada, Hanson, Hembree, Peng, Shrestha, Stewart, Wright and Jasieniuk2013) reported that multiple points of origin were suspected for GR C. canadensis biotypes in California, with regional but not statewide spread. In spite of the potential for long-distance dispersal by C. canadensis, both of these studies concluded that the GR biotypes discovered across regions or states could most likely trace their origins to commonalities in management practices rather than to shared genetic parentage and subsequent dispersal.
The evolution and discovery of GR weeds in the corn (Zea mays L.) and soybean [Glycine max (L.) Merr.] growing region of Canada has frequently followed a pattern mirroring that reported in adjacent states but with a 5- to 10-yr delay. For example, GR C. canadensis was first reported in Delaware in 2001, Ohio in 2002, Pennsylvania in 2003, Michigan in 2007, and Ontario, Canada, in 2010 (Heap Reference Heap2017). From 2010 to 2015, GR C. canadensis has been observed with increasing frequency along a corridor stretching from the first reported case in the most southwesterly corner of Ontario to the northeastern border of the province, just south of Ottawa (Budd Reference Budd2016; Byker et al. Reference Byker, Soltani, Robinson, Tardif, Lawton and Sikkema2013). As might be expected, an increasing number of seed samples of C. canadensis were submitted for resistance testing leading up to and following the discovery of glyphosate resistance in this species in 2010. Many of these samples and their associated collection information have been retained in the seed collections of the University of Guelph and Agriculture and Agri-Food Canada. While these samples certainly do not represent an exhaustive survey of the genetic diversity of C. canadensis from southwestern Ontario, they do offer a unique window into the relatedness of GR accessions and the glyphosate-susceptible (GS) accessions that predate the evolution of resistance. The primary objectives of this study were to: (1) characterize the genetic variation of C. canadensis accessions from the province of Ontario using simple sequence repeat (SSR) markers and (2) investigate the molecular mechanism(s) conferring glyphosate resistance in these accessions.
Materials and Methods
Seed Sources
Conyza canadensis seeds were obtained from the collections of the University of Guelph and Agriculture and Agri-Food Canada’s Harrow Research and Development Centre. These collections primarily contained C. canadensis entries representative of southwestern Ontario from the late 1990s to 2015 (Supplementary Table S1). A total of 98 C. canadensis accessions were included in this study, some of which have been described in previous publications such as Smisek et al. (Reference Smisek, Doucet, Jones and Weaver1998), Weaver et al. (Reference Weaver, Downs and Neufeld2004), and Byker et al. (Reference Byker, Soltani, Robinson, Tardif, Lawton and Sikkema2013). Also included in this study were accessions that we considered distinct from the main collection in terms of chronology and geography. For example, an entry from the University of Guelph collection (i.e., Cc93) was sourced from a Dominion of Canada, Department of Agriculture seed catalog that was composed of vials containing example of economically important crop and weed seeds for use by seed merchants and agricultural institutions. Although the catalog itself is undated, it was produced during the period of time when the Honorable Sydney A. Fisher served as minister of agriculture from 1896 to 1911. Other notable accessions encompassed in this study include a GR C. canadensis accession from Michigan and two entries from Delaware: (1) a 2003 sample from the original GR accession described in VanGessel (Reference VanGessel2001) and (2) a subsequent sample from the same location, taken in 2015. It is important to note that the accessions described in this study do not represent a geographically or chronologically standardized survey of C. canadensis in Ontario, nor were the seed of these accessions collected following a uniform protocol (i.e., it is unclear whether seed from an accession represents a sample of one or many plants); rather, they represent what was retained in the seed collections of the University of Guelph and Agriculture and Agri-Food Canada. All available background information for each accession has been included in Supplementary Table S1, including the results of any discriminating glyphosate-dose tests that were carried out at the time of collection.
Growth Conditions and Dose–Response Experiments
As the vast majority of the accessions used in this study had been stored for more than 10 yrs, many did not contain viable seed (i.e., 72 out of 98 accessions). For the 26 accessions for which germinable seed was available, seedlings were propagated under greenhouse conditions with a day/night temperature of 25/15 C and a photoperiod of 16 h. Dose–response assays were conducted when plants had reached the 5-cm-diameter (rosette) growth stage. Rosettes of C. canadensis were sprayed with the potassium salt of glyphosate (Roundup WeatherMax® with Transorb 2 Technology, 540 g ae L−1, Monsanto Canada, 900–One Research Road, Winnipeg, MB, Canada) at doses of 0, 450, 900, 1,800 and 3,600 g ae ha−1. Herbicide was applied to the plants using an automated spray chamber equipped with 8002E even-spray nozzles set to apply at a rate of 333.3 L ha−1 pressurized by CO2. The experimental design for the dose–response trial was a randomized complete block with four replicates. Because of the large number of accessions evaluated in this study and the difficulty of synchronously propagating similarly sized rosettes, not all accessions could be present within an experimental repetition of the dose–response experiment. As a result, some accessions were present in more repetitions than others; however, all accessions were present for a minimum of two repetitions in time, each containing four replicates of all doses (i.e., a minimum of n=8 for each dose evaluated). Plants were assessed 14 d after treatment (DAT) and scored for injury, and the aboveground biomass was harvested, dried to constant moisture, and weighed. Where the maximum reduction in aboveground biomass accumulation was ≥50%, nonlinear regression analysis was used to determine the effect of glyphosate dose on aboveground biomass of C. canadensis accessions. Data from each accession were fit using the PROC NLIN function in SAS (SAS Institute, Cary, NC). Where possible, dose–response data were fit to a log-logistic model (Equation 1; Seefeldt et al. 1995); however, for most GS accessions this equation would not converge. Thus, a three-parameter exponential decay function was fit for these accessions (Equation 2; adapted from Smisek et al. Reference Smisek, Doucet, Jones and Weaver1998).
where D is the aboveground biomass when dose=0 and is bounded at ≤100, C is the lower response limit, ED50 is the herbicide dose that results in a 50% reduction in aboveground biomass, and b is the slope of the curve around ED50.
SSR Genotyping
DNA was extracted from ~20 mg of freeze-dried leaf tissue or ~10 mg of seed using a genomic DNA extraction kit (Nucleospin® Plant II, Macherey-Nagel, Düren, Germany) according to the manufacturer’s protocol. Template DNA concentration and purity were measured with a full-spectrum spectrophotometer (ND-1000, NanoDrop Technologies, Wilmington, DE), and the concentration was standardized to 10 ng/μl for use in PCR reactions with molecular-grade water. SSR markers developed by Abercrombie et al. (2009) and Okada et al. (Reference Okada, Hanson, Hembree, Peng, Shrestha, Stewart, Wright and Jasieniuk2013) were used to genotype the various accessions. Of the initial 14 SSR markers developed in these studies, only 8 were used in our final analysis (Table 1). PCR amplification was performed based on the Schuelke method (Schuelke Reference Schuelke2000), with the final PCR cocktail consisting of the following: 3 μl of 20 % Trehalose, 4.06 μl of molecular-grade H2O, 1.5 μl of 10X PCR buffer, 1.5 μl of 25 mM MgCl2, 1.0 μl of 3 mM dNTP mix, 0.12 μl of 4 μM M13-tailed forward primer, 0.48 μl of reverse primer, 0.48 μl of 4 μM “universal” M13 primer labeled with either 6FAM, VIC, NED, or PET fluorescent dyes (Applied Biosystems, Foster City, CA), 0.4 μl of 2.5 U μl−1 taq polymerase (Sigma Jumpstart™, Sigma-Aldrich, St. Louis, MO), and 3 μl of template DNA for a total reaction volume of 15 μl. Amplification reactions were performed using thermocyclers (Eppendorf MasterCylcer®, Hauppauge, NY) with the following cycling profile: an initial denaturation at 94 C for 5 min followed by a two-step cycling profile, with 30 cycles of 94 C for 30 s, 56 C for 45 s, and 72 C for 45 s followed by 8 cycles of 94 C for 30 s, 53 C for 45 s, and 72 C for 45 s, with a final extension at 72 C for 10 min. Completed PCR products were pool-plexed to combine up to four SSR markers at a time for fragment analysis using a genetic analyzer (ABI 3500, Applied Biosystems). A dye-labeled size standard (GeneScan 500-LIZ, Life Technologies, Burlington, ON, Canada) was used as the internal size standard, and PCR fragment sizes were determined using a DNA analysis software (GeneMarker, Softgenetics, State College, PA) with a local Southern sizing algorithm.
Table 1 Simple sequence repeat markers for Conyza canadensis.

a Abbreviations: F, forward; R, reverse.
The fragment-size values were used to generate a distance matrix based on the simple matching coefficient, and a neighbor-joining (NJ) dendrogram was produced in MEGA 7 (Kumar et al. Reference Kumar, Stecher and Tamura2016) to display the relationships. Principal coordinate analysis (PCoA) and an analysis of molecular variance (AMOVA) were conducted in GenAlEx 6.501 (Peakall and Smouse Reference Peakall and Smouse2012).
Target Gene Sequencing
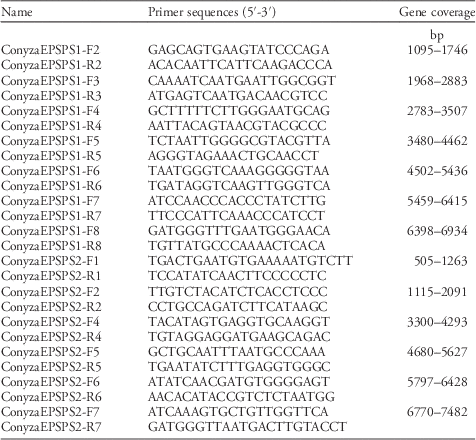
Crop and weed species often have multiple EPSPS gene loci (Filiz and Koc Reference Filiz and Koc2016; Gaines et al. 2013; Garg et al. Reference Garg, Vaid and Tuteja2014; Peng et al. Reference Peng, Lai, Lane, Nageswara-Rao, Okada, Jasieniuk, O’Geen, Kim, Sammons, Rieseberg and Stewart2014). Conyza canadensis has three EPSPS gene loci (i.e., EPSPS1 [AY545666.1], EPSPS2 [AY545667.1], and EPSPS3 [AY545668.1]). Encoded EPSPS1 and 2 proteins show high amino acid sequence similarity and mostly differ in their transit peptide sequence. Introns of EPSPS1 and EPSPS2 genes have low identity. EPSPS3 is also highly homologous to EPSPS2; however, the EPSPS3 gene contains an intron splice site error and does not code for a functional protein (RD Sammons, personal communication). Thus, gene-specific primers were designed based on the Genbank reference sequence for EPSPS1 and EPSPS2 (Table 2). Each PCR reaction consisted of the following: 22 µl of molecular-grade H2O, 9 µl of 20% Trehalose, 5 µl of 10X PCR buffer, 3 µl of 25 mM MgCl2, 1.5 µl of 3 mM dNTP, 2.0 µl each of 10 µM forward and reverse primer, and 0.5 µl of 2.5 U µl−1 Sigma Jumpstart™ taq polymerase. To this mix, 5 µl of 10 ng µl−1 template DNA was added for a total reaction volume of 50 µl. The PCR amplification was as follows: an initial denaturation at 95 C for 2 min, 35 cycles of 95 C for 1 min, variable annealing temperature (depending on primer pair) for 1:30 min, 72 C for 2 min, followed by a final extension at 72 C for 10 min. A 10 µl aliquot of the completed PCR reaction was visualized via 2% agarose gel electrophoresis to evaluate the specificity of the amplified product. A 25-µl aliquot of the completed PCR reaction was purified using a functionalized pipette (Diffinity RapidTip, Chiral Technologies, West Chester, PA) according to manufacturer’s instructions for use as template DNA for bidirectional sequencing of the PCR amplicons. Sanger sequencing was done using premised sequencing reagents (BigDye® Terminator v. 3.1 Cycle Sequencing Kit, Applied Biosystems) and a genetic analyzer (ABI 3500, Applied Biosystems).
Table 2 Primer sequences for EPSPS1 (AY545666.1) and EPSPS2 (AY545667.1).
Sequencing data were analyzed using a DNA analysis software (SeqScape® Software 3, Applied Biosystems) for contig assembly and alignment to the reference sequence for EPSPS1 and EPSPS2. Where polymorphisms were identified, the electropherograms (both forward and reverse) were manually visualized to ensure the quality of base-call exceeded a quality value score greater than 20.
Results and Discussion
Of the 98 C. canadensis accessions examined in this study, 37 were categorized as GR and 44 as GS (Supplementary Table S1). The remaining 17 accessions were not screened at the time of collection, and their susceptibility or resistance to glyphosate is unknown. The classifications of accessions based on the initial discriminating dose tests was validated where possible through dose response and the effective dose that reduced biomass accumulation by 50% (ED50) was calculated. A total of 26 accessions contained viable seed and were reevaluated through dose response. Of these accessions, only one discriminating dose classification was changed (i.e., Cc60 from GS to GR). Based on these results, we contend that the initial discriminating dose results can be relied upon to accurately categorize accessions for which germinable seed was not available.
An NJ dendrogram was generated to visualize the relationships among accessions (Figure 1; Supplementary Table S1). Some of the SSR markers used to generate this dendrogram (Table 1) have been used in previous population genetic studies of C. canadensis notably Yuan et al. (Reference Yuan, Abercrombie, Cao, Halfhill, Zhou, Peng, Hu, Rao, Heck, Larosa, Sammons, Wang, Ranjan, Johnson, Wadl, Scheffler, Rinehart, Trigiano and Stewart2010) and Okada et al. (Reference Okada, Hanson, Hembree, Peng, Shrestha, Stewart, Wright and Jasieniuk2013). In the present study, a total of 52 alleles were observed, with a range of 1 to 13 alleles per marker (Table 1). The NJ dendrogram showed distinct genetic relationships among C. canadensis accessions, particularly with respect to glyphosate susceptibility or resistance. With the exception of five accessions (in Figure 1, counterclockwise from left Cc97, Cc51, Cc60, Cc96, and Cc98), all GR accessions localized to a single cluster on the dendrogram. This cluster was composed of GR accessions collected from southwestern Ontario in 2011 or 2012, with a geographic range of ~315 km.
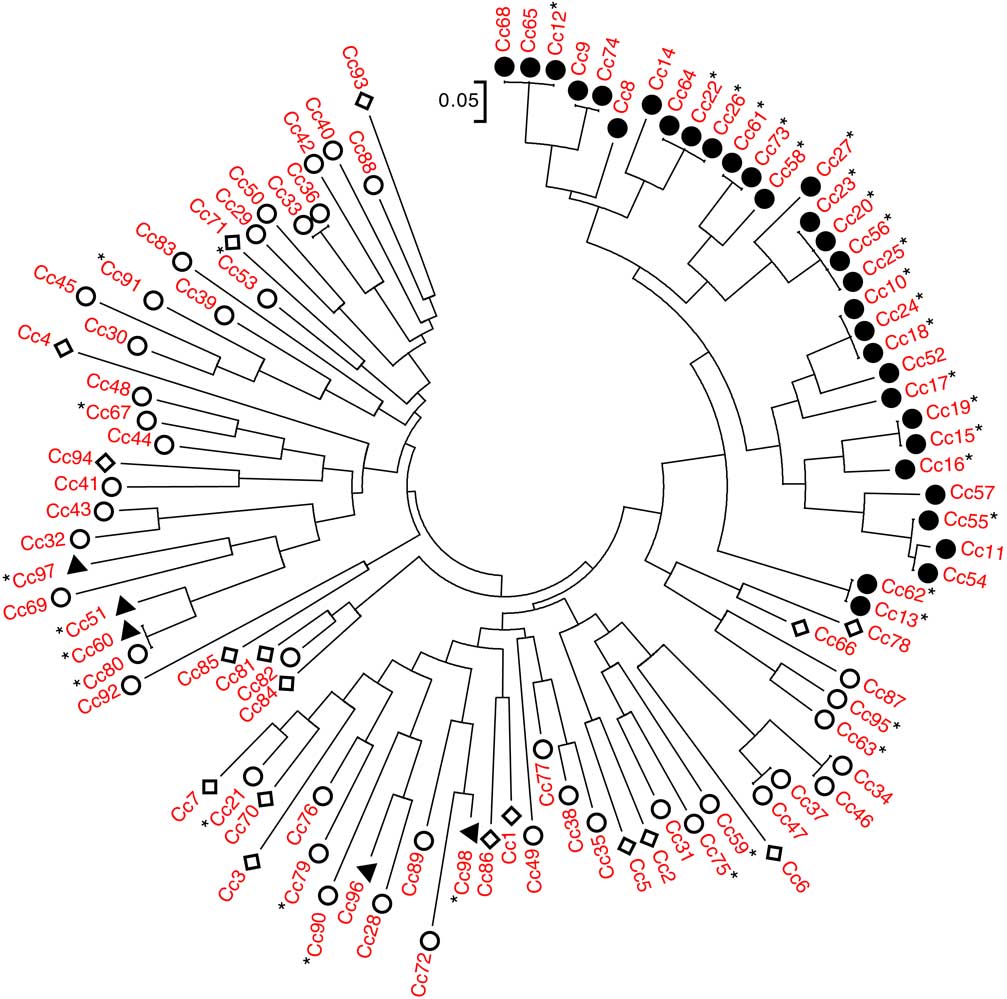
Figure 1 Genetic relationships among the 98 accessions of Conyza canadensis. Entries are characterized as glyphosate resistant (●), non–target site glyphosate resistant (▲), glyphosate susceptible (○), and unknown (◊). Those followed by a star (*) have sequence coverage of exon 2 of EPSPS2. Thus, confirmed target-site resistant entries (i.e., with the Pro-106-Ser substitution) are those preceded by ● and followed by *.
Similar patterns of genetic differentiation among GR and GS C. canadensis accessions were observed in our PCoA (Figure 2). The first two principal coordinate axes accounted for 18% and 9% of the total variance, respectively. In our PCoA, two distinct groups of C. canadensis accessions can be clearly identified: (1) a large group comprising 44 GS accessions, 17 unknowns, and 5 GR accessions (i.e., Cc51, 60, 96, 97, and 98); and (2) a smaller, more compact group comprising 32 GR accessions. Based on these groupings, an AMOVA was conducted (Table 3; Meirmans Reference Meirmans2006, Reference Meirmans2012). This analysis indicated that, while 73% of the total genetic variation was accounted for among the accessions as a whole, 27% was captured by the grouping identified in the PCoA and NJ dendrogram. While this result suggests that there is a significant probability that these groups are genetically distinct subpopulations (FST=0.272, p<0.0001; Meirmans and Hendrick Reference Meirmans and Hedrick2011; Wright Reference Wright1965), the magnitude of this FST value should be interpreted with caution as C. canadensis is a highly selfing species (Charlesworth Reference Charlesworth2003; Hamrick and Godt Reference Hamrick and Godt1996; Smisek Reference Smisek1995).
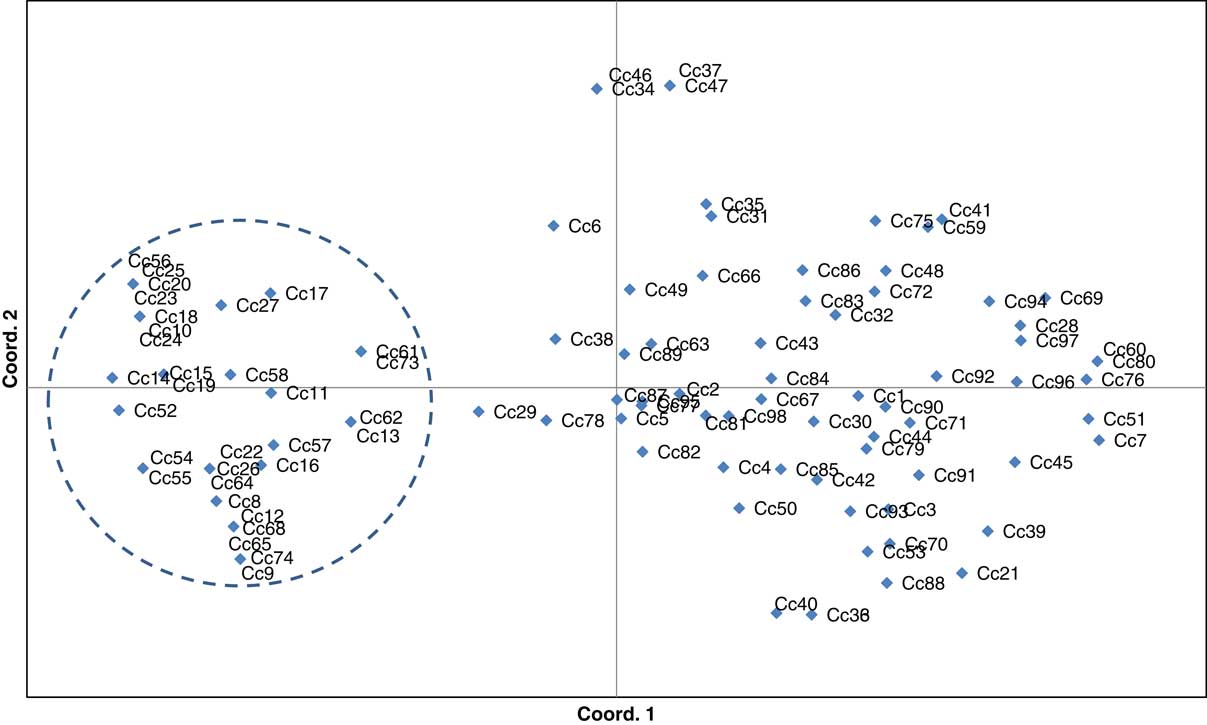
Figure 2 Principal coordinate analysis was constructed using simple sequence repeat Conyza canadensis data. The first two coordinate axes represent 18% and 9% of the observed genetic variation, respectively. Accessions within and outside the dashed line circle represent the population subgroups identified for the purpose of conducting an analysis of molecular variance (see Table 3).
Table 3 Hierarchical analysis of molecular variance based on simple sequence repeat (SSR) data for Conyza canadensis.a , Footnote b
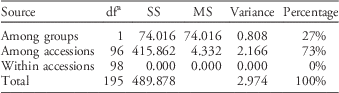
a Abbreviations: df, degrees of freedom; MS, mean square; SS, sum of squares.
b Two groups were identified from principal coordinates analysis and supporting data on glyphosate susceptibility or resistance (Figure 2).
The apparent correlation between subpopulation grouping and glyphosate resistance was further examined through the sequencing of EPSPS1 and EPSPS2. For the purpose of this discussion, we have focused on the DNA sequence of exon 2 in EPSPS1 and EPSPS2, as all single-nucleotide polymorphisms (SNPs) previously reported to confer glyphosate resistance in other species can be found within this region (Sammons and Gaines Reference Sammons and Gaines2014). Complete sequence coverage of exon 2 of EPSPS1 and EPSPS2 was obtained for 29 and 36 of the 98 accessions included in our collection, respectively. A total of 28 accessions had coverage of exon 2 for both EPSPS1 and EPSPS2. For EPSPS1, no differences in the sequence of exon 2 were observed among any of the accessions examined. For EPSPS2, 21 accessions contained an SNP resulting in a proline to serine substitution at position 106 (Pro-106-Ser; Figure 3). Changes in the amino acid at this position have previously been shown to confer glyphosate resistance in several weed species, including goosegrass [Eleusine indica (L.) Gaertn.], tall waterhemp [Amaranthus tuberculatus (Moq.) Sauer], junglerice [Echinochloa colona (L.) Link.], sourgrass [Digitaria insularis (L.) Mez ex Ekman], Italian rygrass (Lolium perenne L. ssp. multiflorum (Lam.) Husnot), and rigid rygrass (Lolium rigidum Gaudin) (Alarcón-Reverte et al. Reference Alarcón-Reverte, García, Urzúa and Fischer2013; Bell et al. Reference Bell, Hager and Tranel2013; de Carvalho et al. Reference de Carvalho, Cruz-Hipolito, González-Torralva, Alves, Christoffoleti and De Prado2011; Jasieniuk et al. Reference Jasieniuk, Ahmad, Sherwood, Firestone, Perez-Jones, Lanini, Mallory-Smith and Stednick2008; Kaundun et al. Reference Kaundun, Zelaya, Dale, Lycett, Carter, Sharples and McIndoe2008; Nandula et al. Reference Nandula, Reddy, Poston, Rimando and Duke2008, Reference Nandula, Ray, Ribeiro, Pan and Reddy2013; Perez-Jones et al. Reference Perez-Jones, Park, Polge, Colquhoun and Mallory-Smith2007; Wakelin and Preston Reference Wakelin and Preston2006). No other SNPs were observed in exon 2 of EPSPS2 for any of the 36 C. canadensis accessions with sequence coverage of this region. When this segregating SNP data from EPSPS2 were overlain onto the NJ dendrogram (Figure 1), it became evident that the groupings observed in this analysis and in the PCoA were directly correlated with the presence or absence of Pro-106-Ser.
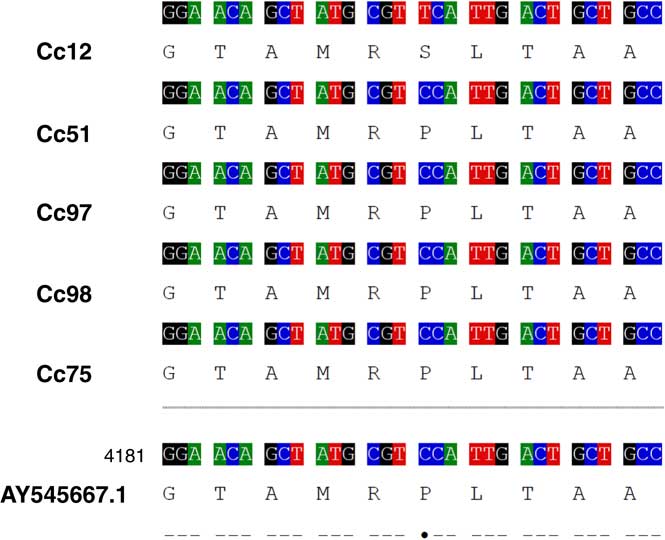
Figure 3 EPSPS2 (gb|AY545667.1|: 4181–4210) sequence of five representative Conyza canadensis accessions: a target-site resistant accession from Ontario (Cc12), non–target site resistant accessions from Ontario, Michigan, and Delaware (Cc51, Cc97, and Cc98, respectively), and a glyphosate-susceptible accession from Ontario (Cc75).
When challenged with glyphosate, accessions from our collection containing Pro-106-Ser (henceforth referred to as target-site resistant [TSR] accessions) displayed markedly higher levels of resistance to glyphosate than GS and known non–target site resistant (NTSR) accessions (e.g., Cc96/Cc98; VanGessel Reference VanGessel2001; Figure 4). For accessions whose resistance status could be classified based on supporting sequence data of EPSPS2, the mean ED50 of GS, NTSR, and TSR accessions were 179, 704, and ≥3,600 g ha−1of glyphosate, respectively (Supplementary Table S1). We chose not to fit an equation to our TSR accessions, because the range of doses tested did not result in a significant (>50%) decline in aboveground biomass relative to the untreated control. Thus, our estimated ED50 for these accessions is greater than or equal to the highest dose evaluated (i.e., 3,600 g ae ha−1). Similar consideration should be given to the range of doses evaluated when interpreting the ED50 values reported for NTSR accessions. For example, the NTSR accessions presented in Figure 4 have ED50 values ranging from 473 to 1,295 g ha−1 (Supplementary Table S1). Based on these values alone, it could be concluded that Cc97 was more resistant than either Cc98 or Cc51, yet at our highest dose, visible injury ratings indicate that Cc97 was controlled, whereas the other two NTSR accessions were still alive (unpublished data). In these particular instances, and indeed in our experiment in general, we acknowledge that there is a lack of sufficiently high doses to force the reductions in aboveground biomass accumulation in TSR and NTSR accessions down to level observed for GS accessions. At present, it is unclear how high a dose would be required to accomplish such a reduction, particularly because TSR accessions have been observed to survive doses as high as 16 times the label rate (ERP, personal observation). Similarly, high levels of resistance have also been observed in horseweed accessions from Ohio and Iowa, with some accessions surviving 20 times the label rate (Beres et al. Reference Beres, Ernst, Snow, Parish, Owen, Ackley and Loux2015).
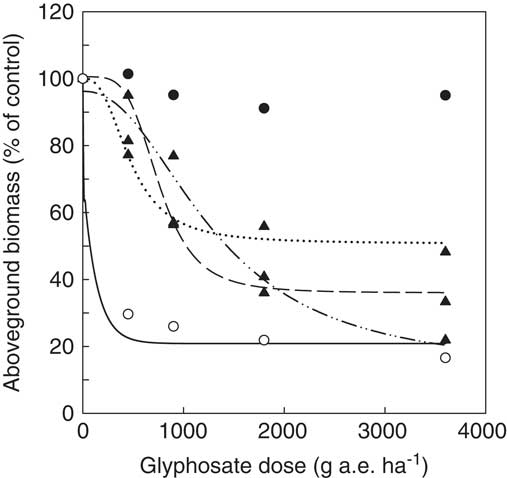
Figure 4 Dose response of five representative Conyza canadensis accessions: a target-site resistant accession from Ontario (Cc12, ●), non–target site resistant accessions from Ontario (Cc51, ▲, ![]() ), Delaware (Cc98, ▲,
), Delaware (Cc98, ▲, ![]() ), and Michigan (Cc97, ▲,
), and Michigan (Cc97, ▲, ![]() ), respectively, and a susceptible accession from Ontario (Cc75, ○,
), respectively, and a susceptible accession from Ontario (Cc75, ○, ![]() ). A four-parameter log-logistic equation (f(x)=C+D−C/1+exp[b(logx)−log(ED
50))]) was fit to Cc51 (C=51, D=100, ED50=473, b=3), Cc98 (C=36, D=100, ED50=763, b=4), and Cc97 (C=13, D=96, ED50=1295, b=2), whereas a three-parameter exponential decay function
). A four-parameter log-logistic equation (f(x)=C+D−C/1+exp[b(logx)−log(ED
50))]) was fit to Cc51 (C=51, D=100, ED50=473, b=3), Cc98 (C=36, D=100, ED50=763, b=4), and Cc97 (C=13, D=96, ED50=1295, b=2), whereas a three-parameter exponential decay function
![]() $\left( {f\left( x \right){\equals}C{\plus}D\,/\,2^{{\left( {x\,/\,ED_{50} \right)}} } \right)$
was fit to Cc75 (C=21, D=100, ED50=134).
$\left( {f\left( x \right){\equals}C{\plus}D\,/\,2^{{\left( {x\,/\,ED_{50} \right)}} } \right)$
was fit to Cc75 (C=21, D=100, ED50=134).
Results of this study are the first to report target-site resistance to glyphosate in C. canadensis. This result is in contrast to all previous studies of glyphosate resistance in C. canadensis which have uniformly identified non–target site resistance mechanisms such as vacuolar sequestration or impaired translocation as the primary mechanisms conferring resistance (Dinelli et al. Reference Dinelli, Marotti, Bonetti, Minelli, Catizone and Barnes2006; Feng et al. Reference Feng, Tran, Chiu, Sammons, Heck and CaJacob2004; Ge et al. Reference Ge, d’Avignon, Ackerman and Sammons2010; Moretti and Hanson Reference Moretti and Hanson2017; Peng et al. Reference Peng, Lai, Lane, Nageswara-Rao, Okada, Jasieniuk, O’Geen, Kim, Sammons, Rieseberg and Stewart2014; Yuan et al. Reference Yuan, Abercrombie, Cao, Halfhill, Zhou, Peng, Hu, Rao, Heck, Larosa, Sammons, Wang, Ranjan, Johnson, Wadl, Scheffler, Rinehart, Trigiano and Stewart2010). Our results also indicate that target-site resistance is the most common mechanism of resistance in Ontario accessions of C. canadensis and that accessions possessing Pro-106-Ser have far greater levels of resistance than NTSR accessions from within or outside this growing region. Why target-site resistance in C. canadensis has become a prevalent mechanism for glyphosate resistance in Ontario alone, more than 10 yr after the initial reports of NTSR accessions in the United States, remains unclear (Heap Reference Heap2017; VanGessel Reference VanGessel2001).
It is important to note that our results cannot exclude the possibility that non–target site resistance mechanisms are also present in the accessions we have characterized as TSR. Indeed, several studies of other weed species have observed target-site mutations acting in concert with non–target site resistance mechanisms (Alarcón-Reverte et al. Reference Alarcón-Reverte, García, Urzúa and Fischer2013; Bostamam et al. Reference Bostamam, Malone, Dolman, Boutsalis and Preston2012; Kaundun et al. Reference Kaundun, Zelaya, Dale, Lycett, Carter, Sharples and McIndoe2008; Nandula et al. Reference Nandula, Ray, Ribeiro, Pan and Reddy2013). In rigid ryegrass, for example, accessions containing only non–target site mechanisms displayed 2- to 4-fold resistance to glyphosate, whereas accessions containing both altered translocation and a target-site mutation displayed 7- to 10-fold resistance (Bostamam et al. Reference Bostamam, Malone, Dolman, Boutsalis and Preston2012). In our study, if we hypothesized that additional non–target site mechanisms were contributing to the levels of resistance observed in TSR accessions, then at a minimum we can conclude that the addition of Pro-106-Ser to non–target site mechanisms, such as those present in Cc51 and Cc60, can significantly enhance the levels of resistance well beyond what has been documented in previous studies of GR horseweed (e.g., Dinelli et al. Reference Dinelli, Marotti, Bonetti, Minelli, Catizone and Barnes2006; VanGessel Reference VanGessel2001).
The first GR C. canadensis accession reported in Canada does not contain Pro-106-Ser (i.e., Cc51; Heap Reference Heap2017). This accession was sampled from the same southwestern Ontario region as the majority of our TSR accessions, only a year earlier (Supplementary Table S1; Supplementary Figure S1). Given this geographic proximity and the sequence of the collections, it seems likely there should be some shared genetic background. Yet when the relationship to the TSR accessions is examined, it is clear that Cc51 is one of the most genetically distant accessions in our entire collection (Figures 1 and 2). For instance, in our NJ dendrogram, the nearest neighbors of Cc51 were an NTSR accession (Cc60) and a GS accession (Cc80), collected approximately 100 and 200 km from Cc51, respectively. In contrast, two GR accessions that cluster with known TSR accessions (i.e., Cc52 and Cc57; Figure 1) were the nearest neighbors of Cc51 both in terms of geography and chronology, having been collected only a year later from sites located less than 10 km from Cc51 (Supplementary Table S1). It is clear from these results that the TSR accessions in our collection not only group together in our NJ dendrogram and PCoA, they do so independently of geographically proximate GS or NTSR accessions (Figures 1 and 2).
While previous studies have had some success in using molecular markers to examine the genetic diversity and relatedness in susceptible and resistant weed populations (Cavan et al. Reference Cavan, Biss and Moss1998; Chandi et al. Reference Chandi, Milla-Lewis, Jordan, York, Burton, Zuleta, Whitaker and Culpepper2013; Lu et al. Reference Lu, Baker and Preston2007; Menchari et al. Reference Menchari, Délye and Le Corre2007; Okada et al. Reference Okada, Hanson, Hembree, Peng, Shrestha, Stewart, Wright and Jasieniuk2013; Riar et al. Reference Riar, Rustgi, Burke, Gill and Yenish2010; Tsuji et al. Reference Tsuji, Fischer, Yoshino, Roel, Hill and Yamasue2003; Yuan et al. Reference Yuan, Abercrombie, Cao, Halfhill, Zhou, Peng, Hu, Rao, Heck, Larosa, Sammons, Wang, Ranjan, Johnson, Wadl, Scheffler, Rinehart, Trigiano and Stewart2010), none have observed the degree of segregation documented in this study or such a strong correlation with an underlying mechanism of resistance. The correlation between the observed grouping and the presence or absence of Pro-106-Ser did not arise from a marker that by chance was linked to the target-site mutation; none of the markers used in our analysis presented an allele that was always and exclusively associated with the TSR group. Several observations can be made of the allelic abundance and diversity among the observed groups that help to explain their divergence: (1) the average number of alleles per marker was greater in the GS/NTSR group than in our TSR group (i.e., 6.375 vs. 2, respectively) and (2) the number of alleles unique to the GS/NTSR group was greater than to the TSR group (i.e., 36 vs. 1 unique alleles, respectively, with 53 shared alleles between groups). Taken together, these observations indicate that our TSR group is defined more so by the reduction in the number and diversity of its alleles relative to the GS/NTSR group than by the presence of unique alleles.
Based on our results, there are several plausible scenarios that could explain the origins of and relationship between our NTSR and TSR accessions. In the simplest case, target-site and non–target site glyphosate resistance mechanisms were selected for independently in the same geographic region from a pool of C. canadensis accessions that possessed at least some shared genetic background. The relative lack of unique alleles in the TSR group suggests that it is unlikely to have arisen via a long-distance dispersal event that would have contributed to the genetic diversity. While this scenario is indeed the simplest explanation, it implies that our TSR accessions possess only a single resistance mechanism and that the high levels of resistance observed in our study can be solely attributed to this target-site mutation. At present this conclusion would run counter to the general consensus that, relative to other herbicide mode of action target-site mutations, mutations in EPSPS endow comparably low levels of resistance (Sammons and Gaines Reference Sammons and Gaines2014).
If we assume that levels of resistance observed in our study are too high to be attributed solely to a target-site mutation, then there must be an additional resistance mechanism or mechanisms acting in our TSR accessions. As discussed earlier and in Sammons and Gaines (Reference Sammons and Gaines2014), there are cases of target-site mutations acting in concert with non–target site mechanisms to provide enhanced levels of glyphosate resistance. It should be noted, however, that the instances in which non–target site resistance and target-site resistance to glyphosate co-occur in the same plant are at present limited to species that are highly outcrossing or obligate outcrossers (i.e., rigid ryegrass and waterhemp; Liu et al. Reference Liu, Davis and Tranel2012; Preston et al. Reference Preston, Wakelin, Dolman, Bostamam and Boutsalis2009). Outcrossing in C. canadensis has been estimated at 4.3% (with a range of 1.2% to 14.5%; Smisek Reference Smisek1995), and based on this comparatively low rate of outcrossing, it seems unlikely that target and non–target site resistance mechanisms would accumulate in C. canadensis through pollen-mediated gene flow. Rather, as our data suggest, the target-site mutation could have been selected for in an accession that had previously evolved some degree of non–target site resistance to glyphosate. The observed groupings in our NJ dendrogram and PCoA could then represent the bottleneck in background genetic variation accompanying the selection for TSR or genetic drift post-selection.
The sequential selection for multiple mechanisms of glyphosate resistance in highly selfing species has been observed previously (Yu et al. Reference Yu, Jalaludin, Han, Chen, Sammons and Powles2015). A double amino acid substitution in goosegrass arose through the sequential selection of target-site mutations Pro-106-Ser and Thr-102-Ile (i.e., the TIPS mutation). In this case, Pro-106-Ser alone provided only moderate resistance to a field dose of glyphosate (i.e., ~30% survival). The authors postulated that the stronger Thr-102-Ile was only selected for in accessions already containing Pro-106-Ser, because the first target-site mutation helped to overcome the deleterious reduction in enzyme kinetics associated with Thr-102-Ile. Ultimately, the combination of the two target-site mutations provided >180-fold resistance to glyphosate, a more than 32-fold increase in resistance when compared with Pro-106-Ser alone.
It is clear from the recent weed science literature that an increasing number of resistance cases can be ascribed to actions of multiple resistance mechanisms (e.g., Alarcón-Reverte et al. Reference Alarcón-Reverte, García, Watson, Abdallah, Sabaté, Hernández, Dayan and Fischer2015; Nandula et al. Reference Nandula, Ray, Ribeiro, Pan and Reddy2013; Yu et al. Reference Yu, Jalaludin, Han, Chen, Sammons and Powles2015). While the results of the present study represent the first report of target site–mediated resistance to glyphosate in C. canadensis, there are several unanswered questions that need to be addressed to better understand the origins and relationship among our TSR and NTSR accessions. Future studies should endeavor to: (1) identify whether other glyphosate resistance mechanisms are present in our TSR accessions, particularly whether impaired translocation or sequestration are present, as these have been previously reported to occur in C. canadensis (Dinelli et al. Reference Dinelli, Marotti, Bonetti, Minelli, Catizone and Barnes2006; Feng et al. Reference Feng, Tran, Chiu, Sammons, Heck and CaJacob2004; Ge et al. Reference Ge, d’Avignon, Ackerman and Sammons2010); (2) determine the mechanism(s) conveying resistance in our NTSR accessions and, with the results from (1), deduce the relationship, if any, between TSR and NTSR accessions; and (3) explore other possible target-site resistance mechanisms in TSR accessions, including determining the copy number and/or expression levels of the three EPSPS loci present in C. canadensis. The fact that Pro-106-Ser occurs in EPSPS2 alone, even though its wild-type mature protein is highly similar to that of EPSPS1, suggests that gene regulation in C. canadensis is such that glyphosate selective pressure has a greater hold on EPSPS2 than EPSPS1.
Acknowledgments
The authors gratefully acknowledge all those who provided seeds of C. canadensis including, but not limited to, Peter Sikkema, Christy Sprague, Mark VanGessel, Mike Cowbrough, Kristen Obeid, and Peter Smith. The authors also would like to thank staff from the Ontario Ministry of Agriculture and Rural Affairs for creating the map of C. canadensis accessions and Sydney Meloche for her contributions toward the dose–response data. Funding for this research was provided by an AAFC A-base grant awarded to ERP (J-000238).
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/wsc.2017.69









