Genetic factors and several postnatal factors such as environmental, social, economic, cultural, psychosocial and behavioural elements are associated with an increased risk for overweight and obesity, and constitute a set of factors that are interrelated and mutually compounding, making overweight/obesity a complex and multifaceted trait( Reference Perez, Garcia and Herrera 1 ). Nevertheless, the factors that best explain the growing number of overweight individuals are related to changes in lifestyle, such as the increased consumption of high-energy processed food, particularly those foods with a high content of SFA and simple carbohydrates, and by increased sedentariness( Reference Kourlaba, Panagiotakos and Mihas 2 ). Currently, there is a growing interest in understanding the genetics of complex traits such as overweight/obesity, which are characterised by a multiplicity of interactions between genetic determinants and conditions associated with lifestyle and the environment.
With the exception of the Y chromosome, studies have shown that all other human chromosomes contain loci associated with the risk for excess weight( Reference Berndt, Gustafsson and Magi 3 , Reference Livingstone, Celis-Morales and Papandonatos 4 ). In various studies using different protocols and different methodologies, more than 430 genes associated with overweight and obesity have been identified( Reference Snyder, Walts and Perusse 5 ). Of all of the genes associated with overweight and obesity, the fat mass and obesity-associated (FTO) gene merits particular mention because it explains the largest amount of genetic variance in obesity traits over the lifespan( Reference Livingstone, Celis-Morales and Papandonatos 4 ). The FTO gene has been associated with energy homoeostasis, participating directly in the control of body fat reserves( Reference Gerken, Girard and Tung 6 ). There is evidence supporting the role of some common genetic variants of the FTO gene in increased food intake and the choice of more energy-dense foods( Reference Moleres, Ochoa and Rendo-Urteaga 7 – Reference Karra, O’Daly and Choudhury 9 ). Currently, studies have shown the effects of components consumed in the diet on gene expression( Reference Qi, Downer and Kilpelainen 8 ). Some dietary components, particularly SFA, are hypothesised to interact with some of the variants of the FTO gene and modify the effect of the association between these genetic variants and obesity in such a way as to compound the obesogenic effect of the FTO gene( Reference Moleres, Ochoa and Rendo-Urteaga 7 , Reference Phillips, Kesse-Guyot and McManus 10 ). However, the mechanisms by which these associations are established remain to be fully clarified.
Previous studies have shown associations of variants of the FTO gene with body weight( Reference Albuquerque, Nobrega and Manco 11 – Reference Zhang, Zhao and Cheng 18 ) but none of these has involved Latin American populations with a high level of miscegenation, as is seen in the north-eastern Brazilian population. Therefore, the objective of the present study was to evaluate the association between SNP in the FTO gene and excess weight in a population of children living in the city of Salvador, Brazil. In addition, the effect of diet as a modifier of this association was assessed in these Brazilian children. The hypothesis was that SNP in the FTO gene are associated with overweight/obesity and that any associations could be modified by dietary intake to modulate the effect of these polymorphisms on excess weight.
Methods
Ethical aspects
The parents or legal guardians of each participating child signed an informed consent form in which the study procedures were described in detail. The study protocol was approved by the internal ethics committee of the Collective Health Institute of the Federal University of Bahia and by the National Research Ethics Council (CONEP) under references 003-05/CEP-ISC and 15.895/2011, respectively.
Study design and population
This is a cross-sectional, population-based study nested within the cohort study Social Changes, Asthma and Allergy in Latin America (SCAALA), which was conducted to evaluate risk factors for asthma in Salvador. The methodology used in that study has been described previously( Reference Barreto, Cunha and Alcantara-Neves 19 ). The city of Salvador has a population of over 2·6 million inhabitants, of whom 80 % consider themselves black or of mixed race( 20 ). The sample consisted of 1445 children who were randomly selected from 20 000 households encompassing a variety of different socio-economic levels and environmental conditions. Of these children, genetic data were available for 1309 children. Following quality control, two children were excluded from the study because data on their obesity/overweight status were missing, and sixty-one were excluded because data on kinship were lacking, leaving 1246 eligible children. After a review of the questionnaires (24-h diet recall), we excluded fifty-five cases because of inconsistent data, and the final sample was made up of 1191 children.
Data collection
Anthropometric data
The participants were weighed on a portable electronic microscale (Filizola®, model E-150/3P, São Paulo, Brazil), and their heights were measured using a portable stadiometer (Leicester Height Measure®; Seca). BMI was used for the diagnosis of anthropometric status, adopting the percentiles for age and sex proposed by the WHO( Reference de Onis, Onyango and Borghi 21 ). Anthropometric status was classified as follows: underweight, <3rd percentile; normal weight, ≥3rd percentile and <85th percentile; overweight, ≥85th percentile and <97th percentile; and obese, ≥97th percentile. The categories of overweight and obese were grouped together; therefore, children classified as having excess weight were those ≥the 85th percentile.
Genotyping
DNA extraction was performed according to the manufacturer’s instructions using the Gentra Puregene Blood Kit. The Qubit fluorometer (Invitrogen) was used for DNA quantification( Reference Costa, Dudbridge and Fiaccone 22 ). All children were genotyped for 458 variants of the FTO gene using the Illumina genotyping platform HumanOmni2.5-8 BeadChip. The Illumina Human Omni 2·5 MV·3 chip delivers comprehensive coverage of both common and rare SNP content from the 1000 Genomes Project (1KGP; minor-allele frequency (MAF)>2·5 %), designed to be maximally informative for diverse world populations. We selected all SNP in the FTO gene (Assembly GRCh37.p13, localised between 53737875 and 54148379 positions) covered by this platform. Genotyping data have been deposited in the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk.ega/), which is hosted by the European Bioinformatics Institute (EBI), under the accession number EGAS00001001245. Imputation was performed only for the variant rs9939609 using the IMPUTE2 package( Reference Howie, Donnelly and Marchini 23 ) on the public panel from 1000 Genomes Project Phase I data ‘version 3’ (ALL.integrated_phase1_SHAPEIT_16-06-14.nomono.integrated_phase1_v3.20101123.snps_indels_svsgenotypes.nomono.haplotypes.gz), which contained 1092 individuals of various ethnicities.
Quality control was performed before conducting the tests for association. All procedures were automated and performed using the PLINK program, version 1.9( Reference Purcell, Neale and Todd-Brown 24 ). To evaluate family structure, kinship coefficients were calculated for every possible pair. A total of sixty-one individuals were removed from the sample because of the relationship determined by kinship coefficients for each possible pair of individuals. This method is implemented in the REAP software (Relatedness Estimation in Admixed Populations)( Reference Thornton, Tang and Hoffmann 25 ). Quality control was conducted in stages to exclude SNP with a genotyping call rate <0·98; deviation from Hardy–Weinberg equilibrium was determined using controls only, with a P-value<0·05 and MAF<1 %( Reference Laurie, Doheny and Mirel 26 ).
Population structure
Principal component analysis (PCA) was conducted to identify different population groups based on the ethnic history/origin of each individual, and correct the population structure( Reference Price, Patterson and Plenge 27 ). Details on the use of PCA to evaluate population stratification can be found in the paper published by Costa et al. in 2015( Reference Costa, Dudbridge and Fiaccone 28 ).
Dietary intake
The 24-h diet-recall method (R24h) was used to determine dietary intake. Parents reported their children’s dietary intake. However, the information given by the children at the time of the interview complemented the information given by their parents. Food consumed in school or at day-care centres was also recorded. The food consumption recorded was converted into energy and macronutrients using DietPro software, version 4.0, 2006( 29 ). Foods that did not form part of the database of this software were added, using information contained in the Brazilian Food Composition Table (TACO)( 30 ), Assessment Table for Food Consumption from Home-Cooking Measurements( Reference Pinheiro, Lacerda and Benzecry 31 ), the ENDEF (Estudo Nacional de Despesas Familiares) food composition table( 32 ) and packaged-food labels. Macronutrient intake was expressed as the percentage of total energy intake.
Statistical analysis
The characteristics of the population were identified by conducting a descriptive analysis. χ 2 Tests were used for categorical variables. The Mann–Whitney U test was used, where appropriate, in order to compare dietary intake variables for ‘overweight’ and ‘not overweight’ groups because of their highly skewed distribution. Logistic regression analysis was used to evaluate the association between genetic variants and excess weight. Each SNP of the FTO gene was analysed separately. Because of the low numbers of AA homozygotes, the genotype was analysed using the dominant-allele model of genetic heritability. The models were adjusted for sex, age, population structure (determined by the first three principal components) and energy intake. These potential confounding variables were selected on the basis of data published in other studies( Reference Qi, Downer and Kilpelainen 33 ). Genomics and proteomics analyses regularly involve the simultaneous testing of hundreds of hypotheses, either on numerical or on categorical data. To correct for the occurrence of false positives, validation tests based on multiple testing correction, such as the Bonferroni and the Benjamini and Hochberg false discovery rate as well as re-sampling techniques (i.e. a permutation-based test) are frequently used. In this paper, we used a permutation test as it is less stringent and has become a widely accepted and recommended approach in studies that involved multiple statistical tests for genetic markers( Reference Belmonte and Yurgelun-Todd 34 ).
Therefore, the empirical P-values were obtained after 50 000 phenotype permutations to limit the occurrence of type-I errors (false-positive results). After the permutation tests, P-values<0·05 were considered statistically significant. Statistical power was estimated with the GAS Power Calculator (http://csg.sph.umich.edu//abecasis/CaTS/)( Reference Skol, Scott and Abecasis 35 ). Linkage disequilibrium values between the evaluated SNP were determined by r 2 in addition to D’ analysis using the Haploview program, version 4( Reference Barrett, Fry and Maller 36 ). Interactions between the FTO genotype and dietary intake, and their effect on overweight/obesity were tested by including the respective interaction terms in the models. All statistical tests were two-tailed, and the significance level was defined as 5 %. All of the statistical analyses were performed using PLINK software, version 1.9( Reference Purcell, Neale and Todd-Brown 24 ).
Results
The eligible study population consisted of 1445 children aged 4–11 years. Of these, 1191 children were included in the analysis, and 11·2 % of these children were overweight/obese. There was a slightly higher percentage of boys than girls (54 v. 46 %) and of children aged 6–7 years (41·8 %). With respect to skin colour, the vast majority of children were reported as being brown or black (91·50 %). The median energy intake was 7045·85 (1928·78–38940·99) kJ. The median intake of carbohydrates, proteins and fat as a percentage of energy content was 61·68 (range: 27·08–87·38), 12·58 (range: 4·28–10·93) and 26·08 (range: 5·28–51·28) %, respectively. The median PUFA:SFA intake was 0·93 (range: 0·02–3·89). There were significant differences in the percentage of energy from carbohydrates (P=0·046) and fat (P=0·037) between the non-overweight/non-obese and overweight/obese groups (Table 1).
Table 1 Characteristics of the population. Salvador, Bahia, Brazil, 2004–2005

* χ 2 Test.
† Mann–Whitney U test.
A genetic analysis, using a logistic regression model adjusted for sex, age, population structure and energy intake, was conducted to evaluate the association between the FTO gene variants and excess weight. As shown in Table 2, ten SNP in introns 1, 7 and 8 of the FTO gene were associated (P<0·05) with excess weight in the population evaluated (online Supplementary Fig. S1). The top SNP associated with excess weight were rs115530394 and rs75066479 for the dominant genetic model. The results of all 400 SNP are presented (online Supplementary Table S1). In addition, a regional association plot was produced using LocusZoom( Reference Pruim, Welch and Sanna 37 ) for better understanding of the genomic context (online Supplementary Fig. S2).
Table 2 Logistic regression between FTO variants and excess weight. Salvador, Bahia, Brazil, 2004–2005
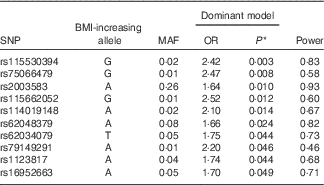
FTO, fat mass and obesity; MAF, minor-allele frequency.
* P=permutation test (50000 permutations). Dominant model.
A polymorphism associated with a particular trait is not necessarily a causal mutation; rather, it may be in linkage disequilibrium with a functional variant. For this reason, an initial investigation was conducted to determine whether SNP associated with excess weight in the Brazilian population capture a single signal or whether they are independently associated with this condition. Of all FTO polymorphisms significantly associated with excess weight, only two pairs were in strong linkage disequilibrium. One pair is located in intron 7 (rs115530394/rs114019148) and the other in intron 8 (rs75066479/rs115662052). In addition, we observed low linkage disequilibrium between the variants rs62048379 and rs9939609 (r 2 0·004 and D’=0·17; r 2 0·004 and D’=0·17). We included rs9939609 in the analysis and indicated that it was imputed (Fig. 1).

Fig. 1 Linkage disequilibrium (LD) of the SNP associated with excess weight. Salvador, Bahia, Brazil, 2004–2005.
Interaction between FTO variants and dietary intake on overweight/obesity
An analysis was conducted of the effect of dietary intake on the association between FTO gene variants and excess weight with the objective of evaluating the modifying effect of diet on this association. Interactions between the FTO genotype and dietary intake, and the effect on overweight/obesity were tested by including the respective interaction terms in the models. For these analyses, the percentage contribution of energy obtained from carbohydrates, proteins, total fat and MUFA was dichotomised according to whether this percentage was above or below the median, as was the ratio between PUFA:SFA.
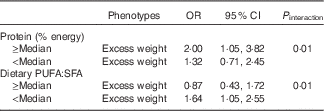
Table 3 shows the results of the multivariate logistic regression analysis conducted to evaluate the association between FTO gene variants and overweight/obesity according to dietary intake. An interaction was found between protein intake and the FTO gene variant rs62048379 (P interaction=0·01) in relation to overweight/obesity. The risk allele for the FTO gene (A allele) was positively associated with overweight/obesity in individuals whose percentage energy intake from protein was above the median (OR 2·00; 95 % CI 1·05, 3·82). An interaction was also found between fat intake (the ratio between PUFA:SFA) and the FTO gene variant rs62048379 (P interaction=0·01) in relation to overweight and obesity. The risk allele for the FTO gene (A allele) for the variant rs62048379 was positively associated with overweight/obesity in individuals whose PUFA:SFA ratio was below the median (OR 1·63; 95 % CI 1·05, 2·55). These data are complemented by the data in the online Supplementary Table S2. Analyses of the remaining FTO gene variants associated with overweight/obesity as a function of dietary intake are shown in the online Supplementary Table S3.
Table 3 Association between polymorphisms (rs62048379) and phenotypes according to dietary intake. Salvador, Bahia, Brazil, 2004–2005 (Odds ratios and 95 % confidence intervals)
Discussion
The present study investigated the possible associations between variants of the FTO gene and overweight/obesity in children in Salvador, Bahia, Brazil. The Brazilian population is considered to be highly admixed. According to Table 1, 91·5 % of the SCAALA population declares itself to be black or brown. Actually, Lima-Costa et al. show that 50 % of the genetic composition in the Salvador-SCAALA cohort is of African origin( Reference Lima-Costa, Rodrigues and Barreto 38 ). Following the appropriate adjustments, SNP that were significantly associated with overweight/obesity were rs115530394, rs75066479, rs115662052, rs114019148, rs62048379, rs62034079, rs79149291, rs1123817, rs16952663 and rs2003583. Variants of the FTO gene influencing obesity have been studied in children in Europe( Reference Albuquerque, Nobrega and Manco 11 ), China( Reference Fang, Li and Du 12 ) and Japan( Reference Okuda, Hinoda and Okayama 13 ), as well as in African American( Reference Bollepalli, Dolan and Deka 39 ) and Brazilian children (including the rs9939609 SNP)( Reference da Silva, Zandona and Vitolo 14 , Reference Lourenco, Qi and Willett 15 ). However, other studies conducted with children of the same age group in Mexico( Reference Mejia-Benitez, Klunder-Klunder and Yengo 16 ), Greece( Reference Scott, Bailey and Moran 17 ) and China( Reference Zhang, Zhao and Cheng 18 ) failed to identify any significant association. The association between overweight/obesity and different variants of the FTO gene in different populations may depend on ethnicity or differences in gene–environment interactions( Reference Tan, Zhu and He 40 ).
Previous studies identified associations between obesity and polymorphisms in introns 1( Reference Foraita, Gunther and Gwozdz 41 , Reference Qi, Downer and Kilpelainen 33 ) and 8( Reference Adeyemo, Chen and Zhou 42 ) of the FTO gene. Those previous findings, together with the present results, suggest the occurrence of variants with a regulatory potential in different regions of the FTO gene. In the present analysis, variants in introns 1, 7 and 8 of the FTO gene were identified as being associated with overweight/obesity. Here, most SNP associated with the trait in question are located in intron 8. This indicates that, in addition to intron 1, intron 8 may constitute a second site of FTO variants related to overweight/obesity. In a study of African American individuals, an association was found between seven SNP from the FTO gene (rs708262, rs11076017, rs16952725, rs9932411, rs7191513, rs2689269, rs16952987 and rs8057044) and overweight/obesity. Some of these SNP are also situated in intron 8( Reference Adeyemo, Chen and Zhou 42 ). These results reinforce the suggestion that there are similarities between genetic factors that predispose individuals of African American descent to obesity and predisposing genetic factors in the Brazilian population, particularly in the admixed population of Bahia( Reference Kehdy, Gouveia and Machado 43 ).
The possible associations between FTO gene variants and overweight/obesity were evaluated in accordance with the composition of the diet. The results of studies investigating the interaction between dietary factors and FTO gene variants on overweight/obesity have generated conflicting results regarding potential interactions( Reference Sonestedt, Roos and Gullberg 44 , Reference Corella, Arnett and Tucker 45 , Reference Phillips, Kesse-Guyot and McManus 10 ). In the present study, an interaction was identified between the percentage of energy intake from proteins and obesity risk linked to the rs62048379 SNP (P-interaction=0·01). An interaction was also found between fat intake (PUFA:SFA ratio) and obesity risk linked to the rs62048379 SNP (P interaction=0·01). The OR that reflect the magnitude of the association between FTO gene variants and overweight/obesity are clearly heterogeneous in this study. Therefore, in individuals whose percentage energy intake from proteins was above the median, the presence of the risk allele (A) for the rs62048379 variant increased the prevalence of excess weight by 2·0 times compared with when the allele was absent. These findings are consistent with the results of a meta-analysis conducted by Qi et al.( Reference Qi, Downer and Kilpelainen 8 ). Those authors showed that a lower dietary protein intake attenuated the association between the FTO genotype and adiposity in children and adolescents. Other studies, however, reported contradictory results( Reference Huang, Qi and Li 46 , Reference Zhang, Qi and Zhang 47 ). Studies suggest that FTO may influence body composition through cellular sensing of amino acids( Reference Gulati, Cheung and Antrobus 48 , Reference Cheung, Gulati and O’Rahilly 49 ). Given the increasing evidence supporting the role of FTO in protein metabolism and body composition, future investigations on this topic may help clarify the mechanisms underlying the observed interaction between the FTO variant and protein intake and its effect on overweight/obesity.
The presence of the allele (A) for the rs62048379 variant increased the prevalence of excess weight by 1·63 times in those individuals whose PUFA:SFA ratio was below the median compared with when this allele is absent. A novel finding in this study was that low dietary PUFA:SFA intake ratio accentuated obesity risk in the A allele carriers but not in the TT homozygote carriers in this population, suggesting that genetic predisposition to obesity may be modulated by dietary SFA intake. The findings of the present study reinforce the results of other studies in which the authors emphasise that diets based on unsaturated fats( Reference Corella, Arnett and Tucker 45 , Reference Moleres, Rendo-Urteaga and Zulet 50 ) may attenuate the obesogenic effect of FTO on the expression of the obesity phenotype. Studies have indicated a possible ‘anti-obesity’ effect attributable to PUFA. This may be due to their greater oxidative rates when compared with SFA( Reference Krishnan and Cooper 51 ). Several investigators have shown that fatty acid oxidation increases directly with a concomitant increase in PUFA:SFA intake( Reference Phillips, Kesse-Guyot and McManus 10 , Reference Westerterp, Smeets and Lejeune 52 ). The mechanisms allowing dietary SFA to interact with FTO are unknown and require further investigation.
The large amount of genome-wide association studies leave no doubt that FTO genotypes in humans are linked to obesity. Recently, researchers have begun to elucidate the underlying pathophysiology behind the mechanism by which this gene may act to increase the risk for obesity( Reference Claussnitzer, Dankel and Kim 53 , Reference Smemo, Tena and Kim 54 ). In fact, very new data obtained using integrated analyses of long-distance regulation, chromatin–chromatin interactions, topologically associated domains, chromatin modifications, gene expression and transgenic animal models have shown that intronic variants within FTO may interact with the promoter region of IRX3/5, affecting its expression( Reference Claussnitzer, Dankel and Kim 53 , Reference Smemo, Tena and Kim 54 ). Genetic variants in FTO and IRX3 were already described to be in high linkage disequilibrium( Reference Srivastava, Mittal and Prakash 55 ). IRX3 was originally found to be related to the early stage of neural development( Reference Bellefroid, Kobbe and Gruss 56 ) and has already been described to be up-regulated in the hypothalamus, which plays a role in food intake or appetite regulation( Reference Smemo, Tena and Kim 54 ). In this way, IRX3 may act as a functional long-range target of obesity-associated variants within the FTO gene and might drive weight gain and the development of overweight and obesity in carriers of common SNP in the FTO gene( Reference Smemo, Tena and Kim 54 ). Further studies are needed to better explore this point.
Our study was based on an analysis of cross-sectional data, which limits the ability to investigate causality. Another limitation of the present study is related to limited statistical power due to the relatively small cohort size. Thus, we chose the dominant heritability model instead of the additive or recessive model in order to improve the power for statistical analysis. Even so, studies with larger sample sizes are needed to support the findings of the present study. In addition, we were unable to examine other adiposity proxies but were limited to the consideration of BMI, which cannot distinguish body composition and does not provide any indication of body fat distribution. The R24h used in the present study to investigate dietary intake may also represent another limitation. Although it is a rapid, relatively inexpensive and easily applied method, the success of its use depends on the respondent’s memory and requires a well-trained investigator to obtain accurate estimations of the portions consumed. However, this method supplies reliable estimates of the mean dietary intake of a population, even when applied only once, as long as the designated methodology is followed and analytical resources are appropriate( Reference Willett and Stampfer 57 ). Most of the children included in our analysis are predominantly of African descent, and it is unknown whether our results can be generalised to other ethnic groups. In contrast, the strengths of this study include the genotyping of various SNP along the FTO gene (400 SNP). An additional strength is the use of a permutation test rather than other methods to test for associations and control for family-wise error rate (FWER), such as Bonferroni and false discovery ratio (FDR). The Bonferroni correction assumes complete independence between markers, but markers in proximity to each other or in low linkage disequilibrium are not completely independent, which makes this approach overly stringent here. The FDR avoids the problem of dependence of tests, but it offers weak control for FWER. It permits a number of false discoveries; therefore, uncertainty remains about the accuracy of each significant result. Permutation analyses provide much stronger control for FWER than the FDR( Reference Groppe, Urbach and Kutas 58 ). As in all approaches, permutation has its weaknesses. Power decreases when there is a very large number of tests. It is most accurate for simple analyses, unless the sample size is large.
In summary, the majority of SNP associated with obesity in this study are located in intron 8, rather than in intron 1, suggesting that there may be many genetic variants that have not yet been reported in the literature that affect obesity in humans. Furthermore, the association between FTO gene variants and excess body weight can be modified by dietary characteristics, particularly by fatty acid distribution and lower dieta protein intake in children, offering new insights into the interrelationships between FTO genetic variants, dietary intake and obesity.
Acknowledgements
This study was funded by the following sources: Funding of Studies and Projects (FINEP, Ministry of Science and Technology, Brazil), the Brazilian National Research Council (CNPq) and The Wellcome Trust, UK, Ref 072405/Z/03/Z.
M. V., G. N. O. C., P. O. and R. d. C. R.-S. participated in statistical analysis, interpretation of results and writing of the manuscript. M. L. B., L. C. R., S. M. A. d. M. participated in the study design, data collection, interpretation of results and revision of the manuscript. C. A. F., N. M. A.-N., A. R. and R. L. F. participated in the interpretation of results and revision of the manuscript. All authors read and approved the final manuscript.
The authors declare that there are no conflicts of interest.
Supplementary material
For supplementary material/s referred to in this article, please visit https://doi.org/10.1017/S0007114517001386







