Introduction
Goosegrass [Eleusine indica (L.) Gaertn.] is a troublesome summer annual weed in turfgrass. Preemergent herbicides are often preferred for control as postemergent herbicide options are limited (McCullough et al. Reference McCullough, Yu and Gomez de Barreda2013). Dinitroaniline and pyridine are two families of mitotic-inhibiting herbicides that are commonly used for preemergent control of annual weeds in turfgrass systems (Breeden et al. Reference Breeden, Brosnan, Breeden, Vargas, Eichberger, Tresch and Laforest2017; McElroy and Martins Reference McElroy and Martins2013). Mitotic-inhibiting herbicides are classified by the Herbicide Resistance Action Committee (HRAC) as Group 3 herbicides with the dinitroaniline family, including the active ingredients prodiamine, pendimethalin, and oryzalin, and the pyridine family, including the active ingredient dithiopyr. Mitotic-inhibiting herbicides inhibit the synthesis of microtubules, which affects root and shoot development (McElroy and Martins Reference McElroy and Martins2013). Dinitroanilines bind directly to the tubulin protein, which prevents the polymerization of the tubulin protein dimer (Vaughn and Lehnen Reference Vaughn and Lehnen1991). Dithiopyr has been reported to inhibit the polymerization of microtubules by binding to microtubule-associated proteins (MAPs) that aid in the stabilization of microtubules; however, research associated with MAPs inhibition by dithiopyr is limited (Vaughn and Lehnen Reference Vaughn and Lehnen1991). All mitotic-inhibiting herbicides result in swollen root tips, as cells at the growing points are unable to divide properly (Vaughn and Lehnen Reference Vaughn and Lehnen1991).
Eleusine indica was first reported as having resistance to mitotic-inhibiting herbicides in 1984 (Mudge et al. Reference Mudge, Gossett and Murphy1984). This resistant biotype was able to withstand up to 6 times the recommended rate of the dinitroaniline herbicide trifluralin (Mudge et al. Reference Mudge, Gossett and Murphy1984). In 1990, two different biotypes of E. indica in South Carolina were reported as having varying levels of resistance to dinitroaniline herbicides (Vaughn et al. Reference Vaughn, Vaughan and Gossett1990). The first biotype had reported levels of resistance that ranged from 1,000- to 10,000-fold, and the second biotype had reported levels of resistance at roughly 50-fold (Vaughn et al. Reference Vaughn, Vaughan and Gossett1990). It was not until 1998 that mutations conferring resistance to mitotic-inhibiting herbicides were reported in E. indica that revealed target-site resistance to dinitroaniline herbicides with mutations found at two different locations on the gene that encodes the α-tubulin protein (Anthony et al. Reference Anthony, Waldin, Ray, Bright and Hussey1998; Yamamoto et al. Reference Yamamoto, Zeng and Baird1998). A high level of resistance was reported for a mutation at the 239 position from threonine to isoleucine (Thr-239-Ile) in E. indica (Anthony et al. Reference Anthony, Waldin, Ray, Bright and Hussey1998). The Thr-239-Ile mutation has also been reported to cause resistance to dinitroaniline herbicides in rigid ryegrass (Lolium rigidum Gaudin) (Fleet et al. Reference Fleet, Malone, Preston and Gill2018). When compared with a population that had a Thr-239-Ile mutation and a susceptible population, a population with a mutation at position 268 from methionine to threonine (Met-268-Thr) was reported to confer intermediate level of resistance to dinitroaniline herbicides in E. indica (Yamamoto et al. Reference Yamamoto, Zeng and Baird1998). To the best of our knowledge, resistance to dithiopyr has not been reported in E. indica, nor has cross-resistance been reported from biotypes that are resistant to dinitroaniline herbicides (Heap Reference Heap2021).
There have been other mutations on the α-tubulin gene that have been confirmed to confer resistance to mitotic-inhibiting herbicides on weed species other than E. indica. A mutation at position 136 from leucine to phenylalanine (Leu-136-Phe) that conferred resistance to dinitroaniline herbicides was discovered in green foxtail [Setaria viridis (L.) P. Beauv.] (Délye et al. Reference Délye, Menchari, Michel and Darmency2004). In 2011, two new mutations were reported on the α-tubulin gene of shortawn foxtail (Alopecurus aequalis Sobol.), one at position 125 that resulted in an amino acid substitution from leucine to methionine (Leu-125-Met) and one at position 202 that resulted in an amino acid substitution from valine to phenylalanine (Val-202-Phe), both of which conferred resistance to the dinitroaniline herbicide trifluralin (Hashim et al. Reference Hashim, Jan, Sunohara, Hachinohe, Ohdan and Matsumoto2012). In 2018, two novel mutations were discovered in L. rigidum, Arg-243-Met and Arg-243-Lys, that conferred resistance to dinitroaniline herbicides (Chu et al. Reference Chu, Chen, Nyporko, Han, Yu and Powles2018). These target-site mutations have not been reported in E. indica.
A suspected dithiopyr-resistant E. indica population was collected in a bermudagrass [Cynodon dactylon (L.) Pers.] putting green on a golf course in Alabama in 2018. The suspected resistant population was noticed after 10 yr of applying mitotic-inhibiting herbicides. An initial evaluation revealed that recommended field rates of dithiopyr (560 g ai ha−1) and prodiamine (735 g ai ha−1) failed to control the resistant population. The objectives of this research were: (1) determine the resistance levels to dithiopyr, prodiamine, pendimethalin, and oryzalin in the suspected resistant population; and (2) to determine whether target-site mutations exist. We hypothesized that the suspected resistant E. indica population was resistant to dithiopyr as well as cross-resistant to dinitroaniline herbicides.
Materials and Methods
Twenty plants from a suspected resistant E. indica population were collected in 2018 from a golf course in Alabama that requested to remain anonymous. The plants were transplanted into a flat filled with potting medium (Scotts Miracle-Gro Products, Marysville, OH), fertilized (28-6-16 Miracle-Gro Water-Soluble All-Purpose Plant Food, Scotts Miracle-Gro Products) biweekly with approximately 25 kg N ha−1, and irrigated three times daily by an elevated misting system to maintain healthy growth. Plants from a known susceptible population were also collected from the Alabama Agricultural Experiment Station, Plant Breeding Unit in Tallassee, Alabama, commonly referred to as ‘PBU’ in past research (McElroy et al. Reference McElroy, Head, Wehtje and Spak2017). Both populations were propagated for seed. Seeds were collected from these plants and combined dried for 48 h and then stored at 4 C for future use.
Dose–Response Screen
Experiments were conducted in a glasshouse environment from May to June 2020. No supplemental light was provided, and the greenhouse conditions were 30 ± 2 C throughout the experiment. The trials were conducted at the same time but were separated by space. Dose–response experiments were carried out on the suspected resistant population (R) and the susceptible population (S) using dithiopyr (Dimension 2EW®, Dow AgroSciences, Indianapolis, IN), prodiamine (Barricade® 4FL, Syngenta Crop Protection, Greensboro, NC), pendimethalin (Pre-M® AquaCap™, LESCO, Cleveland, OH), and oryzalin (Surflan® A.S., United Phosphorus, King of Prussia, PA). Doses for each herbicide were: 0 (untreated control), 0.1, 1.0, 10.0, 100.0, 1,000.0, and 10,000.0 g ai ha−1. Pots with a volume of 230 cm3 were filled with Marvyn loamy sand (fine-loamy, kaolinitic, thermic Typic Kanhapludults) with 0.8% organic matter and pH of 6.3. Twenty seeds were planted in each pot, and soil was added (∼2 mm) to the top to just cover the seeds. The pots were irrigated three times daily by an elevated misting system. The pots were fertilized after planting as previously described every 2 wk for the remainder of the experiment. The pots were then sprayed the following day. The herbicides were applied using a handheld CO2-pressurized sprayer that was equipped with TeeJet® TP 8002 flat-fan nozzles (TeeJet Technologies, Glendale Heights, IL). The sprayer was calibrated to apply 280 L ha−1 at 206 kPa. The experiment was arranged as a completely randomized block design with three replications per treatment, and the experiment was repeated in time. At 6 wk after treatment the number of emerged seedlings and the aboveground fresh biomass per pot were recorded.
α-Tubulin Sequencing
Transcriptome sequencing was conducted to determine whether there were any target-site mutations known to confer resistance to mitotic-inhibiting herbicides. To start the RNA extraction, 100 mg of leaf tissue was collected and ground using a bead mill homogenizer (Omni International, Kennesaw, GA). An RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) was used to extract RNA following the manufacturer’s instructions. DNA digestion was performed using a Turbo DNA-free™ Kit (Applied Biosystems, Foster City, CA) to eliminate any genomic DNA content in the samples. RNA concentration and quality were checked on a NanoDrop 2000 (ThermoFisher Scientific, Waltham, MA) and Qubit 2.0 Fluorometer (ThermoFisher Scientific). RNA integrity was determined using electrophoresis in 2% (3 g / 150 mL) agarose gel. RNA was sequenced via an Illumina NovaSeq 6000 instrument (Novogene, Beijing, China) yielding approximately 45 million 150-bp, paired-end reads. Data were assembled using Trinity (https://github.com/trinityrnaseq), and the resulting assembly was annotated using Trinotate (https://trinotate.github.io). Putative α-tubulin contigs were extracted based on BLASTN (NCBI, https://blast.ncbi.nlm.nih.gov/Blast.cgi) annotation utilizing an E. indica α-tubulin contig (AJ005599). α-Tubulin contigs were aligned with closely related grass species (XM_025935602 and XM_004981865), including other E. indica sequences, using CLUSTAL W+ (www.clustal.org). Illumina sequencing reads for the R population were submitted to NCBI under BioProject number PRJNA725201, and the transcriptome data for the susceptible population can be found on NCBI under BioProject number PRJNA259633.
Data Analysis
Herbicide dose–response data were subjected to ANOVA at a significance level of P < 0.05 using the PROC GLM procedure in SAS v. 9.4 (SAS Institute, Cary, NC) to test for significance (P < 0.05) of populations, herbicide rate, and runs with seedling emergence and aboveground biomass variables. Means and standard errors were generated using the LSMEANS procedure in SAS v. 9.4 (SAS Institute). Means and standard errors were graphed, and I50 values, or the rate needed to reduce emergence or biomass by 50%, were generated using Prism v. 9.0.0 (GraphPad Software, San Diego, CA). Before modeling, the eight rates for each herbicide (including the non-treated) were transformed to log rates with the non-treated set to −3 to maintain equal spacing between treatments. The log-transformed rates were −3, −2, −1, 0, 1, 2, 3, 4, corresponding to 0, 0.01, 0.1, 1.0, 10.0, 100.0, 1,000.0, 10,000.0 g ai ha−1 for each herbicide. I50 values, R2 values, and the other parameters for each equation were calculated for all populations and herbicides based on regression models. If not inherent to the model, I50 values were calculated for each equation. Seedling emergence and biomass reduction data for dithiopyr, prodiamine, pendimethalin, and oryzalin were converted to percent relative to the non-treated.
Three models were used to analyze the data. Seedling emergence control ratings for both populations in response to dithiopyr, prodiamine, and pendimethalin and biomass reduction control ratings for both populations in response to dithiopyr and pendimethalin and the R population in response to prodiamine were modeled using a log(dose) versus response curve equation,
where Y is seedling emergence (%) or biomass reduction (%), X is the log rate of the herbicide, Top and Bottom are plateaus, and logI50 is the log rate of the herbicide that is needed to reduce the seedling emergence by 50%. A line equation was used to model seedling emergence control rating for both populations in response to oryzalin and to model the biomass reduction control rating for the S population in response to oryzalin. The line equation is:
where Y is either seedling emergence (%) or biomass reduction (%), Slope is the rate of reduction, and Y Intercept is where the line intersects the y axis. An exponential plateau equation was used to model biomass reduction for the S population in response to both prodiamine and oryzalin. The equation used was:
where Y is the biomass reduction (%), Y M is the maximum, Y 0 is the starting point, k is the rate constant, and X is the log rate of the herbicide.
Results and Discussion
Our initial hypothesis was that the suspected R population would be resistant to dithiopyr and cross-resistant to the tested dinitroaniline herbicides, because initial screenings failed to control it. Experimental results show that the R population responded differently than the S population to all herbicides. The R population showed higher emergence than the S population when treated with dithiopyr, prodiamine, pendimethalin, or oryzalin (Figure 1). The R population also accumulated more aboveground biomass than the S population when treated with dithiopyr, prodiamine, pendimethalin, or oryzalin (Figure 2). Based on the I50 values for both seedling emergence and biomass reduction, the R population is highly resistant to dithiopyr, pendimethalin, and oryzalin (Table 1). However, the resistance level to prodiamine was low (Table 1).

Figure 1. Seedling emergence response of resistant (R) and susceptible (S) Eleusine indica populations to increasing rates of dithiopyr (A), prodiamine (B), pendimethalin (C), and oryzalin (D). Seedling emergence is relative to the nontreated. Vertical bars are standard errors of individual means. Model components and I50 values are presented in Table 1.
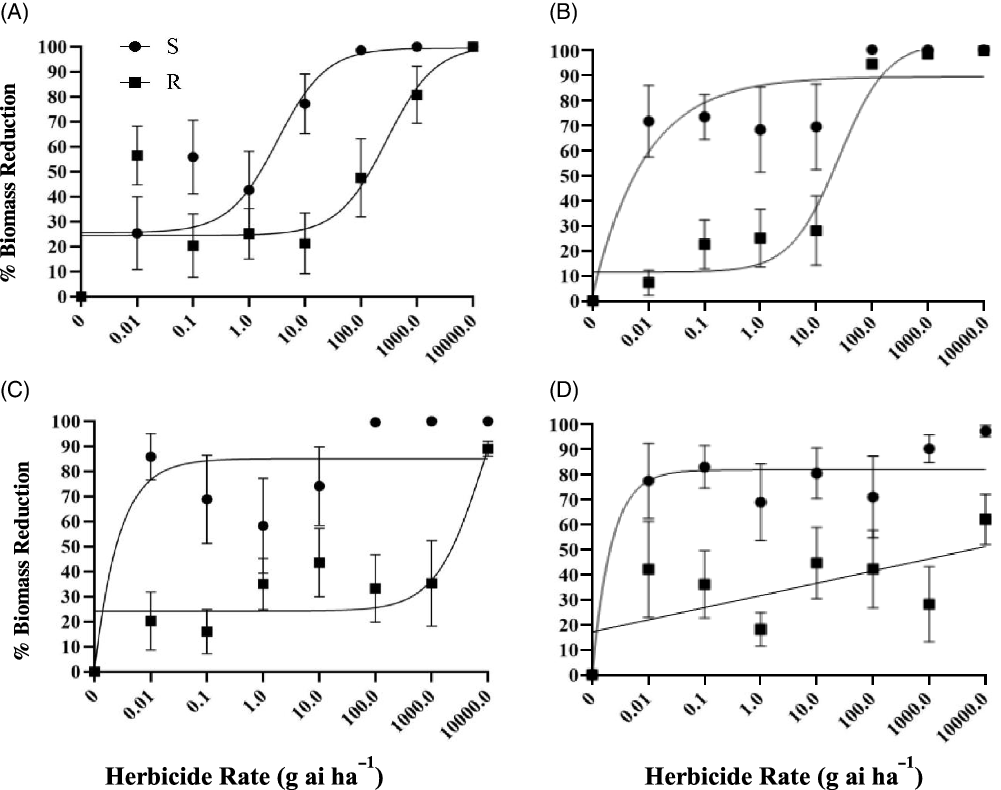
Figure 2. Biomass reduction response of resistant (R) and susceptible (S) Eleusine indica populations to increasing rates of dithiopyr (A), prodiamine (B), pendimethalin (C), and oryzalin (D). Biomass reduction is relative to the non-treated. Vertical bars are standard errors of individual means. Model components and I50 values are presented in Table 1.
Table 1. Rate at which 50% of seedling emergence and biomass is reduced and the parameter values for resistant (R) and susceptible (S) Eleusine indica populations for dithiopyr, prodiamine, pendimethalin, and oryzalin. I50 is in g ha−1.

The I50 values indicated that higher concentrations of dithiopyr, pendimethalin, and oryzalin were needed to control the R population, while a lower concentration was needed for prodiamine. For seedling emergence, I50 values for dithiopyr, pendimethalin, and oryzalin were 919.2, 7,640, and >10,000 g ha−1, respectively, while the prodiamine I50 value was at least 10-fold less at 73.60 g ha−1. The level of resistance was also lower for prodiamine than the other herbicides when compared with the S population’s I50 value. The R population’s resistance to prodiamine was 4.7-fold greater than the S population’s resistance, while the R population’s resistance to dithiopyr, pendimethalin, and oryzalin was 54.1-, >100-, and >100-fold greater, respectively. A similar trend was observed with biomass reduction with the R population’s I50 values for dithiopyr, pendimethalin, and oryzalin at 285.9, 8,885, and 5,907 g ha−1, respectively, but the R population’s I50 value for prodiamine was less at 25.25 g ha−1. The R population’s I50 value for prodiamine was 7.8-fold greater than the S population’s I50 value, while the R population’s I50 values for dithiopyr, pendimethalin, and oryzalin were 88.4-, >100-, and >100-fold greater than the S population’s I50 values, respectively.
A mutation was discovered at position 136 on the α-tubulin gene that resulted in an amino acid substitution from leucine to phenylalanine (Figure 3). This mutation at position 136 was first reported in S. viridis in 2004 and was confirmed to confer resistance to the dinitroaniline herbicides pendimethalin and trifluralin (Délye et al. Reference Délye, Menchari, Michel and Darmency2004). It was reported again in 2011 in a population of A. aequalis discovered in a wheat (Triticum aestivum L.) field and was confirmed to confer resistance to trifluralin, a dinitroaniline herbicide (Hashim et al. Reference Hashim, Jan, Sunohara, Hachinohe, Ohdan and Matsumoto2012). Until now, this mutation had yet to be reported in E. indica. While it has been previously reported to confer resistance to dinitroaniline herbicides, this mutation, or any other known mutation on the α-tubulin gene, has not been associated with dithiopyr resistance. We are not stating that an α-tubulin mutation is the causal mechanism of dithiopyr resistance at this time, simply that the mutation is present in a dithiopyr-resistant population.

Figure 3. Resistant (R) α-tubulin contig aligned with Eleusine indica, Hall’s panic grass (Panicum hallii Vasey), and foxtail millet [Setaria italica (L.) P. Beauv.] sequences from NCBI. The R population had an amino acid substitution Leu-136-Phe.
Eleusine indica was the first species confirmed as resistant to mitotic-inhibiting herbicides, with the first case of resistance to trifluralin reported in 1973 (Heap Reference Heap2021). Mudge et al. (Reference Mudge, Gossett and Murphy1984) first published on E. indica resistance, reporting that a resistant population was not controlled by 2 times the recommended rate of pendimethalin and oryzalin and 6 times the recommended rate of trifluralin. Anthony et al. (Reference Anthony, Waldin, Ray, Bright and Hussey1998) first identified target-site resistance (TSR) as the causal mechanism of resistance, reporting a 42-fold level of resistance to trifluralin and a 60-fold level of resistance to oryzalin in E. indica. Subsequent TSR, Thr-239-Ile and Met-268-Thr, in E. indica was reported by Yamamoto et al. (Reference Yamamoto, Zeng and Baird1998). Since that time, other target-site mutations have been reported in different grass species. In A. aequalis, a resistant population with mutations at Leu-125-Met and Val-202-Phe resulted in a 5.7-fold level of resistance to trifluralin, while a second resistant population with mutations at Leu-136-Phe and Val-202-Phe resulted in a 30.7-fold level of resistance (Hashim et al. Reference Hashim, Jan, Sunohara, Hachinohe, Ohdan and Matsumoto2012). In L. rigidum, three different target-site mutations at Arg-243-Met, Arg-243-Lys, and Thr-239-Ile resulted in 4-, 8-, and 17-fold levels of resistance to trifluralin, respectively (Chu et al. Reference Chu, Chen, Nyporko, Han, Yu and Powles2018; Fleet et al. Reference Fleet, Malone, Preston and Gill2018).
With the aid of over 40 yr of known resistance, our research demonstrates two points. First, TSR mutations in α-tubulin do not confer equal levels of cross-resistance to all DNA herbicides. Varying levels of cross-resistance have been reported in the past, despite the resistant plants having the same target-site mutation. For example, a Thr-239-Ile mutation has resulted in 42- and 60-fold levels of resistance to trifluralin and oryzalin in E. indica, respectively, and a 17-fold level of resistance in L. rigidum (Anthony et al. Reference Anthony, Waldin, Ray, Bright and Hussey1998; Fleet et al. Reference Fleet, Malone, Preston and Gill2018). Our research demonstrates a similar patten, in that the Leu-136-Phe mutation does not induce equivalent cross-resistance to prodiamine, pendimethalin, or oryzalin. Second, no mutations on the α-tubulin gene have ever been associated with dithiopyr resistance. Dithiopyr is suspected to bind to MAPs, instead of the tubulin protein, that aid in microtubule stability (Vaughn and Lehnen Reference Vaughn and Lehnen1991). In the presence of dithiopyr, MAPs are unable to function properly, resulting in shortened microtubules. However, no functional assay has definitively proven that dithiopyr binds to MAPs. Dithiopyr is suspected to bind to a protein that is 65 kDa, but researchers have not directly identified the protein (Lehnen and Vaughn Reference Lehnen and Vaughn1991). Although dithiopyr does share characteristics with mitotic-inhibiting herbicides, it has a distinctive effect on microtubule organization and stability (Lehnen and Vaughn Reference Lehnen and Vaughn1991). Past research on MAP–dithiopyr interaction are more than 30 yr old, and no modern studies confirming earlier findings are available. In our opinion, the reported MAP–dithiopyr interaction does not preclude possible α-tubulin interaction with dithiopyr. Without more information concerning how dithiopyr interacts with the target protein and with the microtubules, it cannot be determined whether target-site mutations on the α-tubulin gene result in resistance to dithiopyr. So, while we cannot definitively say that the mutation Leu-136-Phe is the causal mechanism of dithiopyr resistance, these findings do warrant further investigation.
The R population is definitely resistant to dithiopyr, pendimethalin, and oryzalin, but is less resistant to prodiamine relative to the other herbicides tested. When the I50 values are compared, the amount of prodiamine needed to control the R population is less compared with the other three herbicides tested. When the I50 value for the R population is compared with the I50 value for the S population, the level of resistance to prodiamine is also much lower than the level of resistance to the other three herbicides tested.
The Leu-136-Phe mutation on α-tubulin was identified and determined to be the cause of resistance to dinitroaniline herbicides. Leu-136-Phe had been previously reported to confer resistance to trifluralin, a dinitroaniline herbicide (Délye et al. Reference Délye, Menchari, Michel and Darmency2004). Cross-resistance to pendimethalin and oryzalin is caused by Leu-136-Phe, but is reduced for prodiamine. While resistance to trifluralin was observed in the two previous experiments involving the target-site mutation Leu-136-Phe, the level of resistance that was reported was much lower than what we observed in our study (Délye et al. Reference Délye, Menchari, Michel and Darmency2004; Hashim et al. Reference Hashim, Jan, Sunohara, Hachinohe, Ohdan and Matsumoto2012). Délye et al. (Reference Délye, Menchari, Michel and Darmency2004) did not publish a level of resistance to the tested dinitroaniline herbicides for the resistant S. viridis population. However, the population that possessed the Leu-136-Phe mutation did have a high survival rate when treated with either pendimethalin or trifluralin. Hashim et al. (Reference Hashim, Jan, Sunohara, Hachinohe, Ohdan and Matsumoto2012) did report a 5.7-fold resistance to trifluralin in the resistant population that contained the Leu-136-Phe mutation; however, this resistant population also possessed a Val-202-Phe mutation. This second mutation could have affected the resistance level to trifluralin; however, without any additional research regarding the Val-202-Phe mutation, the effect on the level of resistance to trifluralin in the resistant population cannot be determined.
While we report variation to mitotic-inhibiting herbicides exists, it is unclear whether there are differences in field response to prodiamine. McCullough et al. (Reference McCullough, Yu and Gomez de Barreda2013) reported on variation in shoot reduction of E. indica resistant to prodiamine. In greenhouse studies, shoot reduction ranged from 4% to 63% at 6 wk after treatment (McCullough et al. Reference McCullough, Yu and Gomez de Barreda2013). Overall control of the resistant population with prodiamine was similar in both greenhouse and field experiments, but there was still a difference with greenhouse and field experiment control levels being <35% and <7%, respectively (McCullough et al. Reference McCullough, Yu and Gomez de Barreda2013). Further research is needed to determine whether the varying resistance to prodiamine seen in our greenhouse dose-response screens translates to variation in control in a field setting.
Acknowledgments
This publication was supported by the Alabama Agricultural Experiment Station and the Hatch Program of the National Institute of Food and Agriculture, U.S. Department of Agriculture. Partial funding for this research was also provided by Bruce Spesard of Bayer Environmental Sciences. Additional funding was provided by the Golf Course Superintendents Association of America’s Environmental Institute for Golf (https://www.eifg.org), the Alabama Turfgrass Research Foundation (https://alturfgrass.org/about-us/research-foundation), and the Alabama Golf Course Superintendents Association (http://alabamachaptergcsaa.com). The authors declared that the research was conducted without any commercial or financial interactions that could be interpreted as likely conflicts of interest.






