Introduction
Pennsylvania smartweed [Persicaria pensylvanica (L.) M. Gómez], a member of the knotweed family (Polygonaceae), is a summer annual broadleaf weed distributed throughout the United States. Persicaria pensylvanica requires wet soil conditions to germinate and can grow up to 1.2m tall and produce 19,000 seeds plant−1 (Sankula and Gianessi Reference Sankula and Gianessi2003). The seed (botanically, achene) can remain viable in undisturbed soil for up to 30 yr and is an excellent source of food for various wildlife such as waterfowl (Sankula and Gianessi Reference Sankula and Gianessi2003; USDA-NRCS 2002). Depending on the plant density (8 to 240 plants 10 m−1 of row), P. pensylvanica can cause yield reductions of up to 62% in soybean [Glycine max (L.) Merr.] (Coble and Ritter Reference Coble and Ritter1978). Askew and Wilcut (Reference Askew and Wilcut2002) reported up to 60% reduction in cotton (Gossypium hirsutum L.) yields at a density of 3 P. pensylvanica plants m−1 of crop row. Potential yield losses of up to 20% were reported in corn (Zea mays L.) (Sankula and Gianessi Reference Sankula and Gianessi2003). Due to its rapid growth rate, high fecundity, complex dormancy, and irregular germination, P. pensylvanica continues to spread to new agricultural production regions (Askew and Wilcut Reference Askew and Wilcut2002; Neubauer Reference Neubauer1971).
Acetolactate synthase (ALS) inhibitors (WSSA Group 2) are one of the most common herbicide sites of action used in many cropping systems. These herbicides ultimately prevent biosynthesis of the branched-chain amino acids valine, leucine, and isoleucine (Umbarger Reference Umbarger1978). There are five different chemical classes of ALS inhibitors: sulfonylureas (SUs), imidazolinones (IMIs), triazolopyrimidines (TPs), pyrimidinylthiobenzoates (PTBs), and sulfonylamino-carbonyl-triazolinones (SCTs). The popularity of ALS-inhibiting herbicides in a wide variety of cropping systems has been attributed to their broad-spectrum weed control, low application rates and mammalian toxicity, and high selectivity (Bellinder et al. Reference Bellinder, Gummesson and Karlsson1994; Park and Mallory-Smith Reference Park and Mallory-Smith2004; Whitcomb Reference Whitcomb1999).
Extensive use of ALS-inhibiting herbicides has resulted in evolution of resistance, more so to these herbicides than to any other site of action (Heap Reference Heap2018). For instance, resistance to ALS inhibitors is now reported in 160 weed species worldwide. The most commonly identified mechanism for resistance to ALS inhibitors is target-site based, due to a missense mutation in the ALS enzyme resulting in herbicide insensitivity (Tranel and Wright Reference Tranel and Wright2002). In weeds, target-site mutations conferring ALS resistance have been reported at eight locations (based on amino acid sequence from Arabidopsis) spread over five different domains (A to E) in the ALS gene (Boutsalis et al. Reference Boutsalis, Karotam and Powles1999; Devine and Eberlein Reference Devine and Eberlein1997).
Persicaria pensylvanica is a common and problematic weed of rice (Oryza sativa L.) production and can reduce both yield and quality of rice (Norsworthy et al. Reference Norsworthy, Burgos, Scott and Smith2006; Smith et al. Reference Smith, Flinchum and Seaman1977). Persicaria pensylvanica was previously reported to be resistant to atrazine (WSSA Group 5) in 1990 in a corn cropping system (Heap Reference Heap2018). However, resistance to ALS inhibitors in this species has not been reported in the United States or elsewhere. ALS-inhibiting herbicides, such as imazethapyr, are routinely used alone or in mixture with other herbicides for controlling smartweeds in imidazolinone-resistant rice (Scott et al. Reference Scott, Barber, Boyd, Selden, Norsworthy and Burgos2018). In the present study, we confirmed resistance to different ALS inhibitors and characterized the underlying mechanism of resistance in a P. pensylvanica biotype from southeast Missouri.
Materials and Methods
Seed Collection and Plant Material Preparation
The resistant (R) plants were collected in 2016 from a field near Hornersville, Dunklin County, southeast Missouri (36.06°N, 90.03°W) after reports of herbicide failure. The field was under a rice–soybean rotation and had a 10-yr history of ALS-inhibitor usage, specifically bispyribac-sodium for P. pensylvanica control in rice. A preliminary screen was conducted at Lonoke Extension Center, Lonoke, AR, on the plants taken from rootstocks obtained from the field. Seeds were collected from the survivors and sent to the Altheimer Laboratory, University of Arkansas, Fayetteville, for further analysis.
Susceptible (S) seed, which was scarified to promote germination, was obtained from River Refuge Seed Company (Brownsville, OR). The R and S P. pensylvanica seeds were planted in plastic trays (25 cm by 55 cm) for germination. One week after germination, seedlings at the 1- to 2-leaf stage were transplanted into 50-well plastic trays (25 cm by 55 cm) filled with potting mix (Sunshine premix No. 1®; Sun Gro Horticulture, Bellevue, WA). Plants were grown in the greenhouse (16-h photoperiod and a 35/25 C day/night temperature) and were fertilized appropriately throughout the study with all-purpose plant food (Miracle-Gro®, Scotts, Marysville, OH).
Whole-Plant Herbicide Dose–Response Experiments
Preliminary experiments were conducted to determine the appropriate dose range for the ALS herbicides used in this study. The following herbicide rates were chosen for R plants: bensulfuron-methyl (Londax®, RiceCo, Memphis, TN) at 1x, 16x, 32x, 64x, 128x, and 256x (x=67.3 g ai ha−1); imazethapyr (Newpath®, BASF, Research Triangle Park, NC) at 0.031x, 0.125x, 0.5x, 1x, 4x, 8x, 16x, and 32x (x=105.8 g ai ha−1), and bispyribac-sodium (Regiment®, Valent USA, Walnut Creek, CA) at 0.062x, 0.5x, 1x, 4x, 8x, 16x, and 32x (x=31.9 g ai ha−1). Susceptible seedlings were treated with the above herbicides at 0.015x, 0.031x, 0.062x, 0125x, 0.25x, 0.5x, 1x, 2x. Fifty seedlings each of R and S P. pensylvanica were included as untreated controls. Once seedlings reached the 3- to 4-leaf stage, they were sprayed with the above herbicides using a research track sprayer equipped with two flat-fan spray nozzles (TeeJet® spray nozzles, Spraying Systems, Wheaton, IL) calibrated to deliver 187 L ha−1 of herbicide solution at 269 kPa, moving 1.6 km h−1. All herbicide treatments included label-recommended adjuvants: crop oil concentrate (Superb® HC, Winfield Solutions, St Paul, MN) at 1% v/v for bensulfuron-methyl, nonionic surfactant (Induce®, Helena Agri-Enterprises, Collierville, TN) at 0.25% v/v for imazethapyr, and a nonionic spray adjuvant and deposition aid (Dyne-A-Pak®, Helena Agri-Enterprises, Collierville, TN) at 2.5% v/v for bispyribac-sodium. Experiments were conducted in two runs with three replications (10 seedlings per replication). Aboveground dry biomass (oven-dried at 65 C) was determined at 3 wk after treatment (WAT).
Statistical Analysis
Aboveground dry biomass (expressed as a percentage of the untreated control) data were analyzed using the ‘drc’ package in R v. 3.1.2 (R Development Core Team 2017). The relationship (P<0.05) between herbicide rate and aboveground dry biomass was established using a four-parameter log-logistic model (Seefeldt et al. Reference Seefeldt, Jensen and Fuerst1995) described as follows:
where Y is the response (dry biomass) expressed as a percentage of the nontreated control, C is the lower limit of Y, D is the upper limit of Y, b is the slope of the curve around GR50 (effective herbicide dose for 50% biomass reduction), and x is the herbicide dose. The resistance index (R/S) was calculated using GR50 values. There was no significant interaction between runs; therefore, data were pooled (P>0.05).
Genomic DNA Isolation
Leaf tissue was collected in liquid nitrogen from eight putative R and two S plants and stored at −80C until further use. Genomic DNA (gDNA) was isolated from the leaves using a modified cetyl trimethylammonium bromide protocol (Doyle and Doyle Reference Doyle and Doyle1987). The quantity and quality of gDNA were determined with a spectrophotometer (NanoDrop 1000, ThermoFisher Scientific, Waltham, MA) and agarose gel electrophoresis.
Gene-Specific Primer Designing
Initial attempts to obtain the full-length ALS gene sequence from the P. pensylvanica using polymerase chain reaction (PCR) were unsuccessful, because no full-length ALS gene sequence from the Polygonaceae family exists in the GenBank database. However, a partial sequence (630 bp) for wild buckwheat [Fallopia convolvulus (L.) Á. Löve] POLCO) (GenBank accession JF826440.1) was available and used in this study (Beckie et al. Reference Beckie, Warwick and Sauder2012). Gene-specific primers from Amaranthus (Diebold et al. Reference Diebold, McNaughton, Lee and Tardif2003) and Russian-thistle (Salsola tragus L.) (Warwick et al. Reference Warwick, Sauder and Beckie2010) were initially used to amplify and sequence the 3′ region of the ALS gene (630 bp) containing the Trp-574 amino acid site, previously associated with ALS-inhibitor resistance in weeds (Table 1). Additionally, primers were designed in the 5′ and 3′ regions of the ALS gene to amplify the remaining nucleic acid sites associated with resistance to ALS inhibitors (Table 1). The forward primer for amplifying the 5′ region of the ALS gene was designed based on the Palmer amaranth (Amaranthus palmeri S. Watson) ALS gene sequence, and the reverse primer was designed from the partial 3′ sequence we obtained. All primers were designed using OligoAnalyzer v. 3.1 (Integrated DNA Technologies, Coralville, IA; IDT SciTools 2017) and checked for specificity using the BLAST tool at the National Center for Biotechnology Information (NCBI, Bethesda, MD).
Table 1 Primers used for amplifying the ALS gene in Persicaria pensylvanica.

Polymerase Chain Reaction and ALS Gene Sequencing
A PCR was performed in a T100 thermal cycler (Bio-Rad, Hercules, CA) for amplification of the 5′ and 3′ regions of the ALS gene. The 50-µl reaction volume consisted of 10 µl of 5x GoTaq Flexi buffer (Promega, Madison, WI), 4 µl of 25 mM MgCl2 (Promega), 1 µl of 10 mM dNTP mix (Promega), 0.25 µl of GoTaq Flexi DNA Polymerase (Promega), 1 µl each of forward and reverse gene-specific primers (Table 1), 2 µl of gDNA and 30.75 µl of nuclease-free water (ThermoFisher Scientific). PCR conditions for amplifying the 5′ region of the ALS gene were 95.0 C for 3 min, 35 cycles of 95.0 C for 30 s, 53.0 C for 30 s, and 72.0 C for 1 min, followed by 72.0 C for 5 min. The PCR conditions for amplifying the 3′ region of the ALS gene were the same, except for the extension step (1.5 min). The PCR products were run on 0.8% agarose gel with 100- and 500-bp markers to confirm amplicon size, purified using the GeneJet PCR purification kit (ThermoFisher Scientific), and quantified with a NanoDrop spectrophotometer. The purified PCR products (25 to 50 ng µl−1) were sent for Sanger DNA sequencing (Genewiz, South Plainfield, NJ), and the resulting R and S ALS gene sequences were aligned using MultAlin to find point mutations (Corpet Reference Corpet1988).
Results and Discussion
Dose–Response Assay
Based on whole-plant dose–response studies, the P. pensylvanica biotype collected from southeast Missouri was found to be resistant to the ALS inhibitors bensulfuron-methyl (SU), imazethapyr (IMI), and bispyribac-sodium (PTB) (Figure 1A–C). The R biotype was highly resistant and exhibited minimal injury to increasing rates of bensulfuron-methyl at 3 WAT (Figure 1A). The bensulfuron-methyl rate that caused 50% reduction in biomass of the S biotype (GR50) was only 2.2 g ai ha−1, whereas for the R biotype, it was 5,127 g ai ha−1 (Table 2). Thus, based on GR50 values, the resistance index (R/S) for bensulfuron-methyl was estimated to be 2,330-fold.
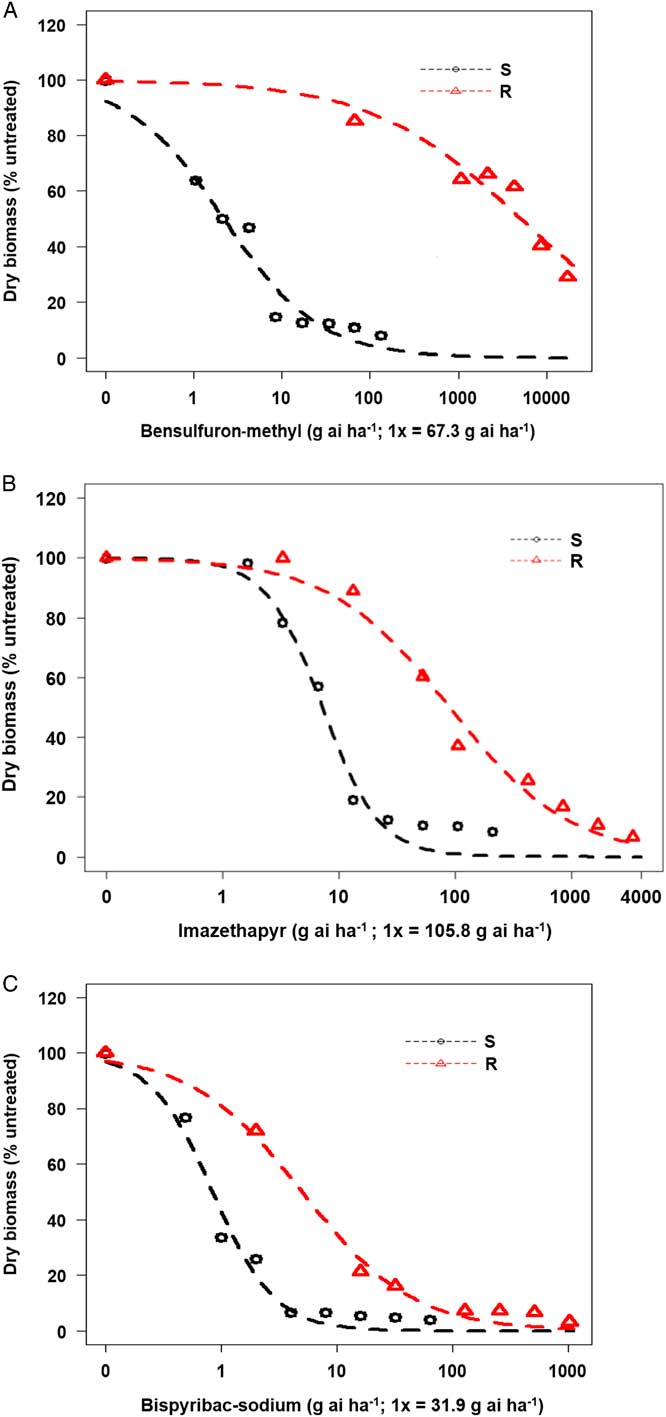
Figure 1 Dose–response assay using four-parameter log logistic model for (A) bensulfuron-methyl, (B) imazethapyr, and (C) bispyribac-sodium herbicides in Persicaria pensylvanica resistant (R) and susceptible (S) biotypes. Each data point is a mean response of 30 plants (10 plants per replication).
Table 2 The effective dose that causes 50% inhibition (GR50) of growth in Persicaria pensylvanica for bensulfuron-methyl, imazethapyr, and bispyribac-sodium.

a Numbers in parenthesis of GR50 values represent SE.
b R/S is the resistance index for each herbicide calculated based on the GR50 values for resistant and susceptible biotypes.
In general, the R biotype survived up to 8x rates of imazethapyr (Figure 1B), showing lack of control at those rates when compared with the S standard. The R plants injured by imazethapyr at 8x rate recovered by 3 WAT. The GR50 for imazethapyr was estimated at 7.2 and 89.3 g ai ha−1, for S and R biotypes, respectively (Table 2). Therefore, the R/S for imazethapyr was estimated to be 12-fold.
Resistance to bispyribac-sodium was the least among the three ALS inhibitors tested (Figure 1C). The GR50 values for bispyribac-sodium were 0.8 and 5.0 g ai ha−1 for S and R P. pensylvanica biotypes, respectively, and the corresponding R/S was only 6-fold (Table 2). The results show that the P. pensylvanica biotype in question is highly resistant to bensulfuron-methyl with a lower level of resistance to imazethapyr and bispyribac-sodium.
Screening for ALS Gene Mutations
The most predominantly identified mechanism of resistance to ALS inhibitors in plants is the presence of amino acid changing point mutations spanning five highly conserved domains of the ALS enzyme (Yu and Powles Reference Yu and Powles2014). The levels of resistance and cross-resistance associated with ALS inhibitors depend on the type of ALS gene mutation and the weed species under investigation (Tranel and Wright Reference Tranel and Wright2002).
In this study, sequencing of the target-site ALS gene from two R plants (out of the eight sequenced) revealed two point mutations, a proline to serine (Pro-197-Ser) and an alanine to serine (Ala-122-Ser), known to cause ALS inhibitor resistance in weeds (Figure 2). Previously, high resistance levels (>340-fold) to the SU herbicides sulfosulfuron and sulfometuron in hare barley [Hordeum murinum L. ssp. leporinum (Link) Arcang.] were attributed to a Pro-197-Ser substitution in the ALS gene (Yu et al. Reference Yu, Nelson, Zheng, Jackson and Powles2007). In contrast, moderate to low levels of resistance to ethametsulfuron (30-fold) and thifensulfuron (~5-fold) were reported in wild mustard (Sinapis arvensis L.) due to the presence of a Pro-197-Ser mutation in the ALS gene (Warwick et al. Reference Warwick, Sauder and Beckie2005). In most cases, point mutations at the Pro-197 position in the ALS gene confer no resistance to IMIs (Devine and Eberlein Reference Devine and Eberlein1997; Devine and Shukla Reference Devine and Shukla2000). It is possible to have the same target-site mutation confer varying levels of herbicide resistance depending on the weed species under study (Sibony and Rubin Reference Sibony and Rubin2003). An ALS mutation in a particular species could be either strong or weak depending on the level of herbicide binding with the target site. For example, both SUs and IMIs bind near, but never directly to, the active site of the ALS enzyme (Yu and Powles Reference Yu and Powles2014).

Figure 2 Two ALS gene mutations conferring ALS-inhibitor resistance in Persicaria pensylvanica. The underlined nucleotides (TCA and TCC) code for the amino acid serine instead of an alanine and proline at two different locations (122 and 197) of the ALS gene. R1, R2, and S refer to two resistant and one susceptible individuals, respectively.
The mutation Ala-122-Ser in the ALS gene reported in our study is an IMI-specific mutation conferring resistance to imazethapyr (Yu and Powles Reference Yu and Powles2014). The target-site mutations in the ALS gene could result in different cross-resistance patterns to ALS inhibitors (Tranel and Wright Reference Tranel and Wright2002; Tranel et al. Reference Tranel, Wright and Heap2018; Wright and Penner Reference Wright and Penner1998). The level of cross-resistance to ALS inhibitors depends on the specific amino acid substituted at the Pro-197 site of the ALS enzyme (Park et al. Reference Park, Kolkman and Mallory-Smith2012). The low-level resistance to bispyribac-sodium (R/S=6) observed in our R population is most likely due to cross-resistance because of the Pro-197-Ser mutation. Tal and Rubin (Reference Tal and Rubin2004) reported cross-resistance to pyrithiobac-sodium and other ALS-inhibiting herbicides in crown daisy [Glebionis coronaria (L.) Cass. ex Spach.] due to substitution of Pro-197 with either serine or threonine.
In this study, only two out of eight R plants sequenced revealed target-site mutations in the ALS gene. The mechanism of ALS-inhibitor resistance in the remaining six plants is unknown. Non–target site resistance mechanisms, such as differences in uptake and translocation or the detoxification of ALS inhibitors, were not investigated in this study. Hence, there is a possibility of having both target-site and non–target site mechanisms existing in the resistant P. pensylvanica biotype.
In summary, this study reports the first documented case, to our knowledge, of field-evolved resistance to ALS inhibitors in P. pensylvanica. We have demonstrated that a biotype collected from southeast Missouri is highly resistant to bensulfuron-methyl and moderately resistant to imazethapyr and bispyribac-sodium. Resistance to these ALS inhibitors will reduce control options for P. pensylvanica in rice under reduced-tillage systems. Use of herbicides with alternative modes of action such as propanil, along with PPO inhibitors such as carfentrazone and acifluorfen, is a good option for controlling ALS-inhibitor resistant P. pensylvanica (Scott et al. Reference Scott, Barber, Boyd, Selden, Norsworthy and Burgos2018).
Acknowledgments
The Arkansas Rice Research and Promotion Board provided funding for this research. The authors express their appreciation to Mallory Everett and Valent USA Corporation for their assistance with this research. No conflicts of interest have been declared.






