Our ‘Cognitive Neurogenetics Project’ started in 2003 with the objective of assessing the influence of APOE and other candidate genes relevant for cognition on normal cognitive aging. A major motivation was to understand neurocognitive mechanisms underlying normal and abnormal cognitive aging, and to identify potential cognitive and brain phenotypes that could be useful for early diagnosis of dementia. Healthy middle-aged and older participants (50–75 years) (N = ~300) were recruited and tested on standard psychometric assays and two experimental cognitive tasks. They also underwent MRI scanning with standard structural (T1-weighted) sequences optimized for automatic processing. Informed consent was obtained to take blood samples and to store biological and behavioral data for 30 years. By 2005–2006, the increasing availability and decreasing cost of genotyping technology made a broader and more general focus on cognitive and imaging genetics potentially amenable. The project was then expanded, increasing the sample size (from ~300 to ~700), expanding the age range (20–80), and performing more expansive cognitive tasks (eight new experimental cognitive tasks), and MRI (diffusion tensor imaging, resting-state fMRI) phenotyping. In 2009, a genome-wide association (GWA) study was performed on the accumulated sample, and the acronym NCNG (Norwegian Cognitive NeuroGenetics) coined and used thereafter to refer to this sample, along with its genetic, imaging, and cognitive data. The central aims of the NCNG are to determine genetic sources of variation in cognitive function, to identify structural and functional brain changes mediating genetic effects, and to study how these relationships are modulated by age. The NCNG is now a relatively large and well-characterized sample of healthy individuals with high-dimensional phenotype and genotype data. This broader data set is better suited to the more general objective of the project, and is also more applicable to other projects within imaging and cognitive genetics, psychiatric genetics, and population genetics. The NCNG is already involved in several collaborations within these fields as a discovery or replications sample, as a healthy control sample, or as a contributor to consortia. The objective of the following presentation is to give researchers information necessary to evaluate possible contributions from NCNG to various multi-sample data analyses. Access to the data for inclusion in multicenter analyses will be reviewed by the NCNG Study Group.
Sample
The sample on which the GWA study was successfully performed consists of 670 persons recruited through newspaper advertisements in the Oslo and Bergen urban areas. All participants were interviewed and probed for past or present neurological or psychiatric diseases known to affect the central nervous system, and for history of substance abuse. Any person with a history of treatment for any of these conditions was excluded from the sample. Participants should have completed basic education with no history of learning deficits; persons who, after initial inclusion, on subsequent testing scored more than one standard deviation (SD) below their age norm on intelligence or memory were excluded. Furthermore, persons with a score on a depression inventory indicating a previously undiagnosed depressive illness were excluded. The participants should be native speakers of Norwegian. All participants gave their informed consent for participation, which included donation of a blood sample, DNA extraction and genotyping, and storage of the remaining blood sample in a biobank. The recruitment procedure resulted in a cognitively normal sample, skewed towards the high functioning intelligence range. Three-to-four-year follow-up data, including the basic test protocol and structural MRI, were collected for available participants above 40 years of age. For further details see Table 1. All participants were also interviewed at each visit according to a ‘Life events questionnaire’, which included questions on health, alcohol consumption, smoking habits, physical exercise, and positive and negative life events (e.g., bereavement through loss of family members or loved ones, loss of work).
TABLE 1 Demographics of the Norwegian Cognitive NeuroGenetics Sample

Follow-up = number of participants that have been re-examined. M = male, F = female, BDI = Beck Depression Inventory, MRI = magnetic resonance imaging.
Genotyping and Population Structure
Due to the original aims of the NCNG Project, genotype data is available for APOE, CHRNA4, and a selection of DNA repair and neuronal plasticity genes for a subset of the sample, as described in Espeseth et al. (Reference Espeseth, Greenwood, Reinvang, Fjell, Walhovd, Westlye, Wehling, Lundervold, Rootwelt and Parasuraman2006), Le Hellard et al. (Reference Le Hellard, Håvik, Espeseth, Breilid, Løvlie, Luciano, Gow, Harris, Starr, Wibrand, Lundervold, Porteous, Bramham, Deary, Reinvang and Steen2009), and Lillenes et al. (Reference Lillenes, Espeseth, Støen, Lundervold, Frye, Rootwelt, Reinvang and Tønjum2011).
For the GWA study, NCNG DNA samples were freshly extracted from blood and genotyped on the Illumina Human610-Quad Beadchip. Quality control was performed with the iterative ‘check.marker’ function in the R package GenABEL (Aulchenko et al., Reference Aulchenko, Ripke, Isaacs and van Duijn2007). Cryptic relatedness was assessed by identity-by-state (ibs), removing one sample from a pair with ‘ibs.threshold’ ≥ .85. Individuals with heterozygosity values greater than two SDs from the sample mean, or with unresolved sex discrepancies, were removed. Population structure was assessed by multidimensional scaling (MDS) analysis (100K random single nucleotide polymorphisms[SNPs]), removing outlying samples with possible recent non-Norwegian ancestry. Finally, SNPs with a call rate < .95, minor allele frequency (MAF) < .01, and Hardy-Weinberg Equilibrium (HWE) exact test P < .001, were excluded. This resulted in a final dataset of 554,225 SNPs genotyped in a homogeneous Norwegian sample of 670 individuals.
The population structure was further assessed by MDS analysis in relation to the original HapMap sample (release 23, downloaded from http://pngu.mgh.harvard.edu/~purcell/plink/res.shtml#hapmap), using approximately 56K overlapping, high-quality SNPs (pairwise r 2 < 0.2, call rate > .99, MAF > .10 and HWE P > .05). The individuals from Bergen and Oslo overlap completely on the MDS plot; the sample is relatively genetically homogenous, clustering at the ‘top’ of the HapMap — CEU sample (CEPH: Utah residents with ancestry from northern and western Europe) (see MDS plot in Figure 1).

FIGURE 1 Multidimensional scaling plot of Norwegian Cognitive NeuroGenetics (NCNG) participants (Oslo and Bergen, Norway) as compared to CEU (Central European, Utah), HCB (Han Chinese, Beijing), JPT (Japanese, Tokyo), and YRI (Yoruba, Ibidan, Nigeria) populations.
Imputed Genotypes
As a member of the ENIGMA consortium (http://enigma.loni.ucla.edu/about/), genotypes were imputed using the ENIGMA protocol available on the web site and described in Stein et al. (Reference Stein, Medland, Arias Vasquez, Hibar, Senstad, Winkler and Thompson2012).
Psychometric Tests and Cognitive Genetics
We used a broad battery of psychometric tests (see Table 2 for the full test protocol), including the Vocabulary and Matrix Reasoning subtests from Wechsler Abbreviated Scale of Intelligence (WASI) (Wechsler, Reference Wechsler1999), to estimate intellectual function (IQ). Furthermore, we included tests of psychomotor speed, attention, and cognitive control functions: the WAIS-R Digit Symbol and the Color-Word Interference Test (CWIT) from the Delis-Kaplan Executive Function System (Delis et al., Reference Delis, Kaplan and Kramer2001), and tests of memory function: the California Verbal Learning Test II (CVLT II) (Delis et al., Reference Delis, Kramer, Kaplan and Ober2000). The CVLT II allows for scoring of several parameters of learning and memory performance, including total learning score, free recall after 5 and 30 min, and recognition of previously learned words measured with recognition hits and false positive responses. The CWIT is an adaptation of the Stroop task (Stroop, Reference Stroop1935), and includes naming of color patches, reading of color words, color-word inhibition, and color-word inhibition/switching. The follow-up protocol included the ‘Mini Mental State’ Examination (MMSE) (Folstein et al., Reference Folstein, Folstein and McHugh1975) to exclude dementia. All participants were interviewed according to a standard protocol about family history of dementia or stroke, and about health conditions. These data have been useful to characterize general cognitive function and health in most NCNG studies, but have also been implemented as the data of interest in several studies. For example, we have been interested in the role of individual differences in cholinergic system function on normal cognition, and in Reinvang et al. (Reference Reinvang, Lundervold, Rootwelt, Wehling and Espeseth2009) we used the psychometric battery to test age by CHRNA4 interactions on cognitive performance. We found that a common polymorphism in CHRNA4 (rs1044396) modulated performance on tests of attention, but not memory, supporting the notion of cognitive specificity of cholinergic pathways reported in earlier pharmacological work in monkeys (Voytko et al., Reference Voytko, Olton, Richardson, Gorman, Tobin and Price1994). Furthermore, on CWIT performance we found stronger effects of the genotype for the oldest participants (70+), suggesting that CHRNA4 might be involved in processes related to accelerated cognitive aging. The same authors (Reinvang, et al., Reference Reinvang, Lundervold, Wehling, Rootwelt and Espeseth2010a) used the longitudinal data set to explore APOE by CHRNA4 interactions on age-related cognitive change, and found that being an APOE ɛ4/CHRNA4 TT carrier was associated with slower and less efficient performance, with steeper decline in speed-related tasks, and in delayed recall in memory tasks; these effects were stronger for the oldest age group (60–79).
TABLE 2 Norwegian Cognitive NeuroGenetics Sample Psychometric Tests
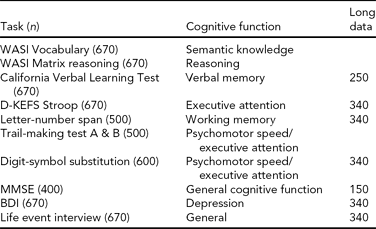
WASI = Wechsler Abbreviated Scale of Intelligence, D-KEFS Stroop = Delis-Kaplan Executive Function System, Color-Word Interference Test, MMSE = Mini-mental State Examination, BDI = Beck Depression Inventory, Long data = longitudinal data.
Le Hellard et al. (Reference Le Hellard, Håvik, Espeseth, Breilid, Løvlie, Luciano, Gow, Harris, Starr, Wibrand, Lundervold, Porteous, Bramham, Deary, Reinvang and Steen2009) used the battery to study the effects of seven genes shown to be up-regulated by infusion of brain derived neurotrophic factor (BDNF) in rat hippocampus (Wibrand et al., Reference Wibrand, Messaoudi, Havik, Steenslid, Lovlie, Steen and Bramham2006). These genes are activity-dependent and are believed to be important for normal learning and memory. We genotyped 271 NCNG participants for tag SNPs covering human orthologs of those genes, and used them in an association analysis. One of those genes, the doublecortin- and calmodulin kinase like 1 (DCLK1) gene, was associated with learning and memory, and with general cognitive performance; similar associations were found in two independent Scottish replication samples.
Recently, Lillenes et al. (Reference Lillenes, Espeseth, Støen, Lundervold, Frye, Rootwelt, Reinvang and Tønjum2011) showed that common polymorphisms in DNA repair genes modulate cognitive aging. The free radical theory of aging proposes that age-dependent accumulation of oxidative damage in DNA and other macromolecules, as well as the genome instability associated with DNA damage, either directly or indirectly leads to aging-associated phenotypic changes at the cellular and organismal levels; the base excision repair (BER) pathway is a primary defense against this accumulation (Harman, Reference Harman1956; Seeberg et al., Reference Seeberg, Eide and Bjørås1995). Cognitive performance is an indicator of general health and also of life expectancy (Thorvaldson et al., 2008). It is therefore possible that individual differences in BER pathway genes could modulate the effect of age on cognitive performance. We genotyped over 700 NCNG participants for 10 non-synonymous SNPs in eight key genes in the BER pathway, and tested their impact on cognitive aging in a combined cross-sectional and longitudinal design. In cross-sectional and longitudinal analyses, we found that the genes hOGG1 and APE1 were associated with effects of age on reasoning ability and attentional performance, but not with level of performance independently of age, suggesting that the effects were specific to aging.
We also used the psychometric data to construct estimates of crystallized and fluid intelligence (g c and g f). Based on a large British sample (N ~3,500), Davies et al. (Reference Davies, Tenesa, Payton, Yang, Harris, Liewald, Ke, Le Hellard, Christoforou, Luciano, McGhee, Houlihan, Gow, Corley, Redmond, Fox, Haggarty, Whalley, McNeill, Goddard, Espeseth, Lundervold, Reinvang, Pickles, Steen, Ollier, Porteous, Horan, Starr, Pendleton, Visscher and Deary2011) were able to show that narrow sense heritability (i.e., the information tagged by SNPs in the GWAS chip through linkage disequilibrium with functional genomic sites) accounted for 40% of the variability in g f and 51% of the variability in g c. Moreover, based on genomic information alone, g c and g f in the NCNG could be significantly predicted (accounting for about 1% of the variability on these estimates).
There are also data from the Scandinavian Odor Identification Test (SOIT) available from ~300 individuals in the NCNG as described in Wehling et al. (Reference Wehling, Nordin, Espeseth, Reinvang and Lundervold2010, Reference Wehling, Nordin, Espeseth, Reinvang and Lundervold2011).
The Genetics of Experimental Cognitive Psychology Tasks
One of the original hypotheses of the project was that cognitive neuroscience paradigms assessing components of attention would show a high degree of sensitivity and specificity for neurogenetic effects. The inclusion of experimental cognitive paradigms is a distinctive aspect of NCNG. Such data are not commonly included in epidemiological studies of cognition, and the lack of standardized paradigms constitutes a barrier to large-scale studies. The importance of developing this type of paradigm for use in clinical or epidemiological research is underscored in the Cognitive Neuroscience Treatment Research to Improve Cognition in Schizophrenia (CNTRICS) initiative (e.g., Barch et al., Reference Barch, Braver, Carter, Poldrack and Robbins2009). In this context the NCNG is a comparatively large sample with a broad range of paradigms, and we will use the GWA study data on NCNG and collaborating replication samples to attempt a genetic parsing of attentional components. Based on the studies by Greenwood and Parasuraman (Reference Greenwood and Parasuraman2003), we selected the Cued Discrimination paradigm as the measure of visual attention in the basic protocol (for details, see Espeseth et al., 2006). This is a version of the extensively studied paradigm originally developed by Posner (Reference Posner1980), where centrally located arrows give a neutral, valid, or invalid cue to the location of peripherally presented visual targets. Data for a replication sample are available from George Mason University (GMU); a collaborative analysis of neurogenetic effects is underway. Espeseth et al. (Reference Espeseth, Greenwood, Reinvang, Fjell, Walhovd, Westlye, Wehling, Lundervold, Rootwelt and Parasuraman2006) reported a replication of the main findings of the study by Greenwood et al. (Reference Greenwood, Sunderland, Friz and Parasuraman2000) on APOE in an independent sample, as well as new data showing interaction between APOE and a nicotinic receptor gene (CHRNA4) in reaction time and white matter volume. We have since obtained data from several additional experimental paradigms in a subset of about 300–500 participants, aiming for a detailed analysis of attention and cognitive control functions (see Table 3 and Appendix 1). We have also acquired pupillometry and fMRI data on the same paradigms within smaller independent samples, to further characterize the physiological mechanisms involved in performing these tasks.
TABLE 3 Norwegian Cognitive NeuroGenetics Sample Experimental Tasks on Attention, Working Memory, and Episodic Memorya
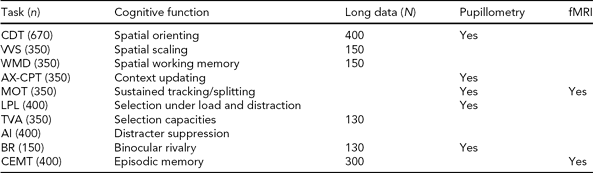
CDT = cued discrimination task, VVS = attentional scaling in visual search, WMD = spatial working memory, AX-CPT = conditional continuous performance, MOT = multiple object tracking, LPL = Lavie perceptual load task, TVA = theory of visual attention, AI = attentional inhibition, BR = binocular rivalry, CEMT = categorized episodic memory task, Long data = longitudinal data.
a See Appendix for brief description of the tasks and representative references.
There are several benefits to detailed sampling of a phenotype with several partly overlapping assays. By using two different cognitive tasks that are believed to share the demand for one latent cognitive function (e.g., attentional effort), while being different on several others, one can use these to test for a conceptual replication of a genetic association while controlling for a larger number of confounding variables. For example, in a study aiming to investigate the role of cholinergic innervation in attentional effort (Sarter et al., Reference Sarter, Gehring and Kozak2006), we combined the Multiple Object Tracking (MOT) (Pylyshyn & Storm, Reference Pylyshyn and Storm1988) and the Lavie Perceptual Load (LPL) (Beck & Lavie, Reference Beck and Lavie2005) paradigms (Espeseth et al., 2010) (see illustration in Figure 2). We found that CHRNA4 genotype modulated performance under high processing load in both tasks, but not under low load. Moreover, since the processing load in the MOT task can be increased parametrically using the same stimulus material, we were able to reveal that the size of the CHRNA4 effect increased linearly up to the capacity limit of approximately four objects. This finding supports the idea that the cholinergic system might mediate increases in attentional effort (Sarter et al., Reference Sarter, Gehring and Kozak2006). However, we did not find any evidence that CHRNA4 modulated processing of irrelevant flankers in the LPL task (a feature that is difficult to test in the MOT task), despite the fact that these distractors have a strong impact on task performance. This suggests that distractor and target processing have at least partly separate neurobiological substrates.
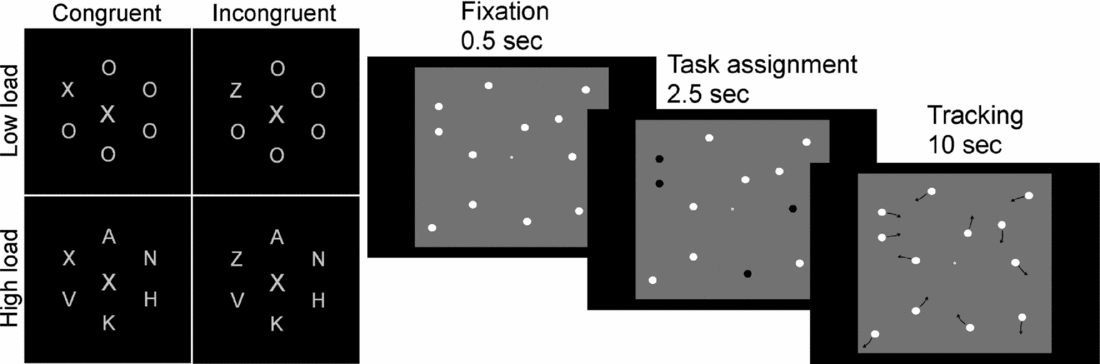
FIGURE 2 Examples of two NCNG (Norwegian Cognitive NeuroGenetics) tasks with application for the current project. The left panel shows four different conditions of a distraction task in which participants are asked to report the letter X or Z in the ‘circle’ of letters while ignoring the central letter. Participants are asked to fixate on the central letter throughout the test period. Performance is typically worse if this letter is dissimilar to the target letter, especially in the low load condition. The right panel shows a multiple object tracking task in which participants are asked to track with their attention only (i.e., no eye movements) from two to six objects that move independently on the screen in an unpredictable manner. Both tasks vary the level of cognitive load and we recently showed that variation in a nicotinic receptor gene modulated performance under high, but not low, load (Espeseth et al., 2010).
Experimental paradigms can also be combined with standard psychometric tests to increase specificity, and perhaps also sensitivity. In a study on the effect of APOE on executive attention and working memory, the AX version of the Continuous Performance Task (AX-CPT) (Braver et al., Reference Braver, Barch, Keys, Carter, Cohen, Kaye, Janowsky, Taylor, Yesavage, Mumenthaler, Jagust and Reed2001) was used concurrently with CWIT, the Letter-Number Span task (LN-S), and CVLT (Reinvang et al., Reference Reinvang, Winjevoll, Rootwelt and Espeseth2010b). APOE was associated with overall performance in LN-S, but this task cannot be used to reveal the cognitive subcomponents involved in the effect. The AX-CPT is designed to delineate subcomponents; the analysis revealed that male APOE ɛ4 carriers had difficulties with suppressing responses to non-targets that followed target cues (i.e., the so-called AY trials).
In a related effort to reveal the computational specificity of attentional functions, we have acquired behavioral data with the ‘CombiTVA’ paradigm (Vangkilde et al., Reference Vangkilde, Bundesen and Coull2011), which is designed to estimate five core parameters of attention: the perceptual threshold, processing speed, visual short-term memory capacity, attentional selectivity, and spatial allocation of attentional weights. Recently, Dyrholm et al. (Reference Dyrholm, Kyllingsbæk, Espeseth and Bundesen2011) published a methodological paper on NCNG data from the ‘CombiTVA’ paradigm, and several genetic association studies involving this paradigm are in progress.
In addition to these experimental paradigms, we have used visual and auditory oddball paradigms in event-related potential studies to reveal effects of APOE and CHRNA4 on electrophysiological brain potentials (Espeseth et al., Reference Espeseth, Endestad, Rootwelt and Reinvang2007, Reference Espeseth, Rootwelt and Reinvang2009, Reference Espeseth, Sneve, Rootwelt and Laeng2010; Reinvang et al. Reference Reinvang, Espeseth and Gjerstad2005).
MRI-Based Imaging
Consenting participants have gone through a standard structural MRI protocol optimized for morphometric analysis in Freesurfer (surfer.nmr.mgh.harvard.edu/fswiki). The Oslo subsample of the NCNG includes 222 participants scanned on a 1.5T Siemens Sonata scanner; 71 of these individuals were re-scanned 3 years later on the same magnet. This scanner gave us little opportunity to include research-oriented sequences outside of the T1-weighted structural scans; thus we decided to move further scanning to a 1.5T Siemens Avanto system at the Department of Radiology, Oslo University Hospital. This scanner enabled us to include sequences for diffusion tensor imaging, resting-state functional MRI, and high resolution T2-weighted 3D fluid attenuated inversion recovery (FLAIR). We also coordinated sequences with another large Norwegian sample scanned on the same magnet. Two hundred and forty-four NCNG individuals have so far completed the Avanto protocol. As part of fMRI projects with cognitive tasks, another 93 NCNG participants have undergone a similar protocol on a 3T Philips Achieva scanner (see Figure 3 for data examples from the Avanto and Achieva magnets). There is some overlap between samples scanned at each of the magnets in Oslo, thus reducing the total number of scans somewhat (see Table 4 for total number of T1-weighted scans in NCNG). In Bergen, more than 100 participants have been scanned twice, of whom a majority also had a third scan, about seven years after the initial one. All scans have been run on a 1.5T GE Signa magnet at the Haraldsplass Deaconess Hospital in Bergen, with no intermediate upgrading of the scanner. All scans were reviewed by a neuroradiologist.
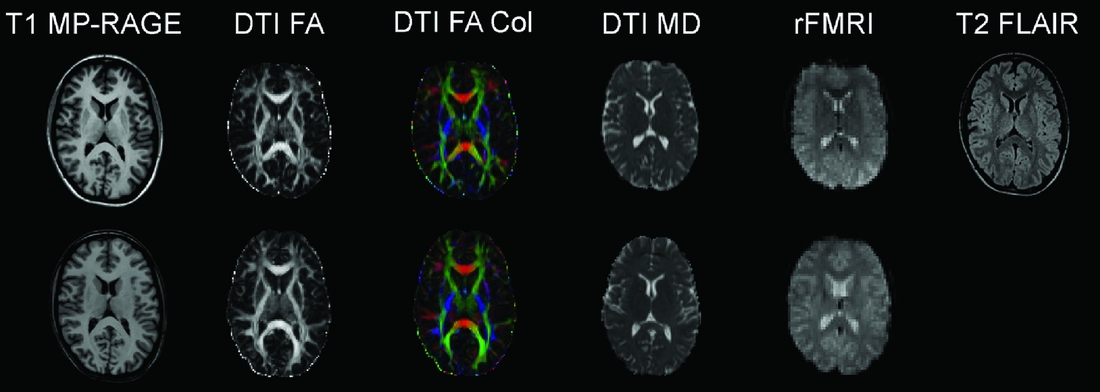
FIGURE 3 Example MRI data from the 1.5T Siemens Avanto scanner (top row) and from the 3T Philips Achieva scanner (bottom row). Both image sets are from the same 23-year-old female participant. T1 MP-RAGE: T1-weighted magnetization prepared rapid gradient-echo, anatomical 3D images; DTI FA: diffusion tensor imaging fractional anisotropy, reflecting the directional coherence of the diffusion pattern where brighter voxels are part of fibers with stronger directional coherence; DTI FA Col: color coded fractional anisotropy map where blue colors denote fibers with superior-inferior orientation, red colors denote fibers with left-right orientation, and green colors denote fibers with anterior-posterior orientation; DTI MD: mean diffusion map of the DTI data; rFMRI: resting-state functional magnetic resonance imaging; T2 FLAIR: T2-weighted fluid attenuated inversion recovery. See Table 4 for protocol details.
TABLE 4 MRI Protocols for the Norwegian Cognitive NeuroGenetics Sample
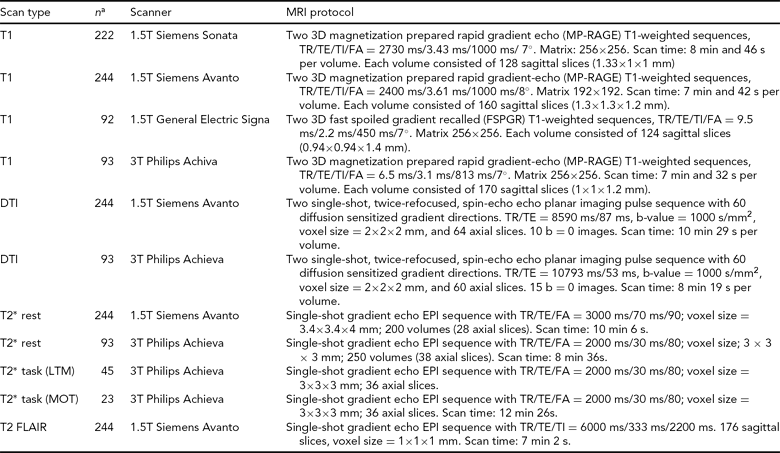
FA = flip angle, TR = repetition time, TE = echo time, TI = inversion time, DTI = diffuse tensor imaging, LTM = long-term memory, MOT = multiple object tracking
a Some overlap between samples: see text for total sample sizes for each scan type.
The Siemens Sonata data set has been included in five multi-sample studies on effects of age on various structural brain properties (Fjell et al., Reference Fjell, Westlye, Amlien, Espeseth, Reinvang, Raz, Agartz, Salat, Greve, Fischl, Dale and Walhovd2009a, Reference Fjell, Westlye, Amlien, Espeseth, Reinvang, Raz, Agartz, Salat, Greve, Fischl, Dale and Walhovd2009b, Reference Fjell, Westlye, Espeseth, Reinvang, Dale, Holland and Walhovd2010; Walhovd et al., Reference Walhovd, Westlye, Amlien, Espeseth, Reinvang, Raz, Agartz, Salat, Greve, Fischl, Dale and Fjell2011; Westlye et al., Reference Westlye, Walhovd, Dale, Espeseth, Reinvang, Raz, Agartz, Greve, Fischl and Fjell2009). Apart from the effects of age on various structures of the brain, an important finding in these studies is that the NCNG data generally show the same results, as do the analyses of other samples from three different countries, involving six different scanners, and slightly different age ranges and recruitment protocols, but where all data sets were analyzed with the same data processing scheme. Analysis of the effect of APOE on white matter volume and cortical thickness has been published by Espeseth et al. (2006, Reference Espeseth, Westlye, Fjell, Walhovd, Rootwelt and Reinvang2008). Further, Espeseth et al. (Reference Espeseth, Westlye, Walhovd, Fjell, Endestad, Rootwelt and Reinvang2012) link APOE-related changes of cortical thickness to variation in attention. Ystad et al. (Reference Ystad, Lundervold, Wehling, Espeseth, Reinvang and Lundervold2009) used both Sonata and GE Signa data to study APOE effects on hippocampal volumes and relations with memory functions. Westlye et al. (Reference Westlye, Lundervold, Rootwelt, Lundervold and Westlye2011) recently published a study revealing effects of APOE on resting-state connectivity based on the GE Signa data. The Neuroinformatics and Image Analysis Laboratory at the Department of Biomedicine, University of Bergen, has provided multi-modal analysis of the Bergen MRI data, and several studies have been published using these methods (e.g., Hodneland et al., Reference Hodneland, Ystad, Haász, Munthe-Kaas and Lundervold2011; Lundervold, Reference Lundervold2010; Ystad et al., Reference Ystad, Eichele, Lundervold and Lundervold2010, Reference Ystad, Hodneland, Adolfsdottir, Haász, Lundervold, Eichele and Lundervold2011). There is, so far, no published multi-modal MRI study based on the Siemens Avanto and Philips Achieva samples. However, the motivation for the multi-modal protocol is similar to the one outlined above on cognitive testing — to enable detailed phenotypic analysis with partly overlapping measures to increase measurement specificity and the ability to identify biologically meaningful associations.
Conclusion and Future Directions
The NCNG sample has several unique features that can make it particularly useful. The NCNG has evolved from a local project to a database with a broad set of data on cognition, brain imaging, and genetics in a life-span sample. The sample originates from a relatively homogenous population, the data have undergone strict procedures of quality control, and the participants have been thoroughly screened, both cognitively and radiologically, to exclude subjects with pathology. Furthermore, the follow-up cognitive testing makes it possible to ensure a healthy sample by excluding from analysis participants with subsequent cognitive decline. This has made NCNG data interesting for inclusion in multi-sample studies of normal aging, and in studies of neurogenetic effects on cognition (Davies et al., Reference Davies, Tenesa, Payton, Yang, Harris, Liewald, Ke, Le Hellard, Christoforou, Luciano, McGhee, Houlihan, Gow, Corley, Redmond, Fox, Haggarty, Whalley, McNeill, Goddard, Espeseth, Lundervold, Reinvang, Pickles, Steen, Ollier, Porteous, Horan, Starr, Pendleton, Visscher and Deary2011), and on brain structure (Stein et al., 2012). Further multi-sample studies are under execution or planning. As outlined above, the phenotyping scheme is very rich and detailed, particularly for experimental cognitive tasks and MRI. It yields a potential for high specificity within these domains, and also within-sample conceptual replications, while at the same time controlling for a number of confounding variables. We have specifically chosen cognitive phenotypes that are based on highly developed models from cognitive neuroscience. The benefit of this strategy is that we can take advantage of knowledge about neural systems that are essential to the execution of cognitive processes involved in performance of these tasks. These factors may contribute to an increased chance of revealing true genetic associations, which in turn may result in a higher rate of replication. Although we have not systematically investigated replication across samples for all the phenotypes, we do have some indications that NCNG data are comparable to those of other large samples. For example, three of the experimental cognitive tasks are identical to tasks used by Parasuraman and Greenwood at GMU, Virginia, USA (see Appendix). The effects of age are similar, and several of the associations with APOE and CHRNA4 are replicated. Preliminary results from several GWA studies on cognitive traits (including both psychometric and experimental data) indicate good replication across several samples from northern Europe and the USA. Analysis of the imaging data is at an early stage, but the pattern seems to be the same.
The fact that results are replicated in similar samples does not guarantee that the results can be generalized to the populations the samples are recruited from. The somewhat skewed cognitive profile of NCNG participants towards higher functioning may limit the representativeness of the results, perhaps especially for the older members of the sample. Patterns of genotype–phenotype association in a high-functioning healthy sample may not be representative for patterns of association in patient samples, potentially limiting the usefulness of NCNG as a control sample in case–control designs. However, the generalizability of NCNG results is an empirical issue, and it is worth mentioning that currently a primary challenge in imaging and cognitive genetics is identification of true replicable signals, which is a prerequisite for generalizability.
Acknowledgments
Many have contributed critically to the development of the NCNG through funding, protocol design, lab resources and data analysis, data collection, and scientific advice. We are particularly grateful to (in alphabetical order) Steinunn Adolfsdottir, Atle Bjørnerud, Claus Bundesen, Sven Cichon, Anders M. Dale, Srdjan Djurovic, Paulina Due-Tønnesen, Tor Endestad, Anders M. Fjell, Pamela M. Greenwood, Per Hoffman, Stefan Herms, Bruno Laeng, Elin Lilleng, Arvid Lundervold, Camilla Middelthon, Athanasia M. Mowinckel, Markus M. Nöthen, Raja Parasuraman, Markus Handal Sneve, Peter Strassegger, Elisabeth Sølsnes, Tone Tønjum, Signe Vangkilde, Kristine B. Walhovd, Eike I. Wehling, and Lars T. Westlye. NCNG has been funded through the Research Council of Norway (including the FUGE program), the National Institutes of Health, the University of Oslo, the University of Bergen, and the Western Norway Regional Health Authority.
Appendix 1
Short Description of Each of the Experimental Cognitive Tasks
1. Cued Discrimination Task (CDT): A cued visual discrimination task developed by Parasuraman et al. (1992), based on the Posner (Reference Posner1980) cued detection task, was used. Participants are given a centrally presented arrow cue to the location at which a letter is subsequently presented. The task is to make a vowel/consonant discrimination on this letter. See Espeseth et al. (Reference Espeseth, Greenwood, Reinvang, Fjell, Walhovd, Westlye, Wehling, Lundervold, Rootwelt and Parasuraman2006) for details on this particular version. Measures endogenous covert visuo-spatial orienting and reorienting, phasic alertness.
2. Attentional Scaling in Visual Search (VVS): A visual search task based on Greenwood et al. (1999). Participants are searching for a pink ‘T’ among 14 distractors while pre-cued by a frame encompassing 1, 3, 9, or 15 letters. Measures the ability or tendency to use prior information on spatial region to adjust the scale of attentional focus to improve letter discrimination in a visual search task involving conditions of both feature and conjunction search.
3. Spatial Working Memory (WMD): A spatial delayed-match-to-sample task involving 1, 2, or 3 locations to be retained over a three-second delay, after which a probe dot appears on one of the relevant locations (match trials), or either 2, 4, or 8° of visual angle away from the nearest relevant location (non-match trials). Experimental details can be found in Greenwood et al. (2005). Measures the capacity and spatial precision of spatial working memory.
4. Conditional Continuous Performance (AX-CPT): A continuous performance task in which the status of the target letter (i.e. ‘X’) is conditional on the identity of the letter presented just before (i.e. ‘A’). ‘AX’ trials are frequent (70%), whereas so-called ‘AY’, ‘BX’, and ‘BY’ trials share the remaining 30% equally. Experimental details can be found in Braver et al (2001) and, for this particular version, in Reinvang et al. (Reference Reinvang, Winjevoll, Rootwelt and Espeseth2010b). Measures working memory updating and conflict resolution (executive attention).
5. Multiple object tracking (MOT): An object tracking task in which 12 objects move independently and unpredictably on the screen, out of which 2–6 objects are targets to be tracked. Based on Pylyshyn & Storm (1988). Experimental details on the current version can be found in Espeseth et al. (2010). Measures sustained attention and maximal attentional tracking capacity, possibly requiring the dividing of attention into separate streams/spotlights.
6. Lavie Perceptual Load Task (LPL): A visual search task based on Beck & Lavie (Reference Beck and Lavie2005). Participants are presented with a circular array of letters containing one target (i.e., ‘X’, or ‘Z’) and five non-targets. The non-targets are all ‘O’ in the low load condition, and letters sharing features with the targets in the high load condition. A response-relevant (i.e., X or Z) or neutral (i.e., ‘L’) distractor is foveally presented, defining congruent, incongruent, and neutral trials. Details of the present version can be found in Espeseth et al. (2010). Measures efficiency of distractor suppression at two different levels of perceptual load.
7. Theory of Visual Attention (TVA): This is the ‘CombiTVA’ task described in detail in Vangkilde et al. (2011). The analysis and parameter estimation is described in Dyrholm et al. (2011). Six letters are presented in a circular array for variable durations where all are targets in ‘whole report’ trials, but only a subset are targets in ‘partial report’ trials. Measures speeds and capacities of critical attentional abilities, such as the lowest presentation time allowable for perceptual awareness, perceptual speed (units per second), capacity of visual short-term memory, the distribution of attentional capacity at different locations of the visual field, and the efficiency of distractor suppression.
8. Attentional Inhibition (AI): Visuo-spatial task based on Stuss et al. (Reference Stuss, Toth, Franchi, Alexander, Tipper and Craik1999). The task involves distractor suppression in three different task settings (i.e., interference, negative priming, and inhibition-of-return) at three different levels of target selection/distractor suppression difficulty.
9. Binocular rivalry: A simple task where participants wear anaglyph spectacles and look at a picture in which a green house is superimposed on a red face (or vice versa) and report perceptual shifts.
10. Categorized Episodic Memory Task (CEMT): Participants view an array of naturalistic images from ten different categories and judge the esthetic attractiveness of each of them. Later, when half the images are presented again intermixed with new ones, participants make old/new judgments combined with a confidence rating. Measures ability of visual episodic memory for naturalistic scenes.









