Serum lipid levels have been associated with cardiovascular diseases, metabolic syndrome, and type II diabetes (Kannel et al., Reference Kannel, Kagan, Revotskie and Stokes1961; Miller & Miller, Reference Miller and Miller1975; Pilia et al., Reference Pilia, Chen, Scuteri, Orru, Albai, Dei, Lai, Usala, Lai, Loi, Mameli, Vacca, Deiana, Olla, Masala, Cao, Najjar, Terracciano, Nedorezov, Sharov, Zonderman, Abecasis, Costa, Lakatta and Schlessinger2006). Variation in lipids levels is highly influenced by heritable factors (Friedlander et al., Reference Friedlander, Austin, Newman, Edwards, Mayer-Davis and King1997), and 95 loci have already been associated with levels of high-density lipoprotein (HDL) cholesterol, low-density lipoprotein (LDL) cholesterol, triglycerides (TG), and total cholesterol (TC) in numerous study samples and replicated in various populations using genome-wide approaches (Aulchenko et al., Reference Aulchenko, Ripatti, Lindqvist, Boomsma, Heid, Pramstaller, Penninx, Janssens, Wilson, Spector, Martin, Pedersen, Kyvik, Kaprio, Hofman, Freimer, Jarvelin, Gyllensten, Campbell, Rudan, Johansson, Marroni, Hayward, Vitart, Jonasson, Pattaro, Wright, Hastie, Pichler, Hicks, Falchi, Willemsen, Hottenga, de Geus, Montgomery, Whitfield, Magnusson, Saharinen, Perola, Silander, Isaacs, Sijbrands, Uitterlinden, Witteman, Oostra, Elliott, Ruokonen, Sabatti, Gieger, Meitinger, Kronenberg, Döring, Wichmann, Smit, McCarthy, van Duijn and Peltonen2008; Kathiresan et al., Reference Kathiresan, Melander, Guiducci, Surti, Burtt, Rieder, Cooper, Roos, Voight, Havulinna, Wahlstrand, Hedner, Corella, Tai, Ordovas, Berglund, Vartiainen, Jousilahti, Hedblad, Taskinen, Newton-Cheh, Salomaa, Peltonen, Groop, Altshuler and Orho-Melander2008; Kooner et al., Reference Kooner, Chambers, Aguilar-Salinas, Hinds, Hyde, Warnes, Gomez Perez, Frazer, Elliott, Scott, Milos, Cox and Thompson2008; Teslovich et al., Reference Teslovich, Musunuru, Smith, Edmondson, Stylianou, Koseki, Pirruccello, Ripatti, Chasman, Willer, Johansen, Fouchier, Isaacs, Peloso, Barbalic, Ricketts, Bis, Aulchenko, Thorleifsson, Feitosa, Chambers, Orho-Melander, Melander, Johnson, Li, Guo, Li, Shin Cho, Jin Go, Jin Kim, Lee, Park, Kim, Sim, Twee-Hee Ong, Croteau-Chonka, Lange, Smith, Song, Hua Zhao, Yuan, Luan, Lamina, Ziegler, Zhang, Zee, Wright, Witteman, Wilson, Willemsen, Wichmann, Whitfield, Waterworth, Wareham, Waeber, Vollenweider, Voight, Vitart, Uitterlinden, Uda, Tuomilehto, Thompson, Tanaka, Surakka, Stringham, Spector, Soranzo, Smit, Sinisalo, Silander, Sijbrands, Scuteri, Scott, Schlessinger, Sanna, Salomaa, Saharinen, Sabatti, Ruokonen, Rudan, Rose, Roberts, Rieder, Psaty, Pramstaller, Pichler, Perola, Penninx, Pedersen, Pattaro, Parker, Pare, Oostra, O'Donnell, Nieminen, Nickerson, Montgomery, Meitinger, McPherson, McCarthy, McArdle, Masson, Martin, Marroni, Mangino, Magnusson, Lucas, Luben, Loos, Lokki, Lettre, Langenberg, Launer, Lakatta, Laaksonen, Kyvik, Kronenberg, König, Khaw, Kaprio, Kaplan, Johansson, Jarvelin, Janssens, Ingelsson, Igl, Kees Hovingh, Hottenga, Hofman, Hicks, Hengstenberg, Heid, Hayward, Havulinna, Hastie, Harris, Haritunians, Hall, Gyllensten, Guiducci, Groop, Gonzalez, Gieger, Freimer, Ferrucci, Erdmann, Elliott, Ejebe, Döring, Dominiczak, Demissie, Deloukas, de Geus, de Faire, Crawford, Collins, Chen, Caulfield, Campbell, Burtt, Bonnycastle, Boomsma, Boekholdt, Bergman, Barroso, Bandinelli, Ballantyne, Assimes, Quertermous, Altshuler, Seielstad, Wong, Tai, Feranil, Kuzawa, Adair, Taylor, Borecki, Gabriel, Wilson, Holm, Thorsteinsdottir, Gudnason, Krauss, Mohlke, Ordovas, Munroe, Kooner, Tall, Hegele, Kastelein, Schadt, Rotter, Boerwinkle, Strachan, Mooser, Stefansson, Reilly, Samani, Schunkert, Cupples, Sandhu, Ridker, Rader, van Duijn, Peltonen, Abecasis, Boehnke and Kathiresan2010; Willer et al., Reference Visscher and Duffy2008). However, the genetic associations affecting blood apolipoproteins have been studied mainly with the candidate gene approach.
Data emerging from epidemiological studies imply that genes interact with environmental factors causing additional variability in lipid levels (Thorn et al., Reference Thorn, Needham, Mattu, Stocks and Galton1998; Visscher & Duffy, Reference Visscher and Duffy2006). In animal and plant models, G × E interactions can be studied by controlling the environmental factors. In human populations, the environment of an individual is highly variable and the individual can choose environments. This makes it difficult to find loci interacting with specified environmental factors. Recently published screen for G × E interactions identified a locus where, depending on the genotype at that locus, the connection between waist-to-hip ratio and total cholesterol differed (Surakka et al., Reference Surakka, Isaacs, Karssen, Laurila, Middelberg, Tikkanen, Ried, Lamina, Mangino, Igl, Hottenga, Lagou, van der Harst, Mateo Leach, Esko, Kutalik, Wainwright, Struchalin, Sarin, Kangas, Viikari, Perola, Rantanen, Petersen, Soininen, Johansson, Soranzo, Heath, Papamarkou, Prokopenko, Tönjes, Kronenberg, Döring, Rivadeneira, Montgomery, Whitfield, Kähönen, Lehtimäki, Freimer, Willemsen, de Geus, Palotie, sandhu, Waterworth, Metspalu, Stumwoll, Uitterlinden, Jula, Navis, Wijmenga, Wolffenbuttel, Taskinen, Ala-Korpela, Kaprio, Kyvik, Boomsma, Pedersen, Gyllensten, Wilson, Rudan, Campbell, Pramstaller, Spector, Witteman, Eriksson, Salomaa, Oostra, Raitakari, Wichmann, Gieger, Järvelin, Martin, Hofman, McCarthy, Peltonen, Van Duijn, Aulchenko and Ripatti2011), but to detect the association, over 40,000 individuals from various population cohorts were used.
The total environmental sensitivity of a locus, without specifying which environmental factors are salient, can be tested in general population samples by comparing the trait variances between different genotype classes in genome-wide association (GWA) studies. For this sensitivity association screening, monozygotic (MZ) twins offer the additional benefit of controlling for possible confounding due to possible epistatic effects. MZ twin-pairs share essentially identical genomes at the sequence level and the differences within pairs; for example, lipid levels reflect environmentally influenced differences. Genetic variation may determine the extent to which environmental differences lead to phenotypic within pair variability (Magnus et al., Reference Magnus, Berg, Borreson and Nance1981). An association with within-MZ-pair trait differences directly indicates gene–environment interaction, where one allele increases or restricts the effect of environmental factors or environmental factors affect the gene expression differently for different alleles. This is seen as elevated or diminished levels of variability in trait values. In a recent methodological paper, the power to detect variability association using either difference between twins or variability in population approaches was evaluated (Visscher & Posthuma, Reference Visscher and Posthuma2010). The study showed that using MZ twins is more powerful than the approach using population variances, when the trait under examination has high within-pair correlations in MZ twin-pairs.
In this study we have screened for genetic “variability” loci where the loci are associated with differences in lipid levels between the two members of a pair in our GWA study in MZ female twins. More specifically, the aim of our study was to search variability genes for HDL, LDL, TC, TG, and apolipoprotein A-I, A-II, B, and E (APOA1, APOA2, APOB, and APOE respectively). For the apolipoprotein data at hand we have additionally screened for apolipoprotein level associations.
Materials and Methods
Study Samples
The Danish, Dutch, Finnish, Italian, Norwegian, and Swedish national twin cohorts, together with St Thomas’ twin cohort in the United Kingdom and an Australian twin cohort, form the basis of the GenomEUtwin project (www.genomeutwin.org), a collection of 800,000 twins (Peltonen & GenomEUtwin, Reference Peltonen2003). From these cohorts, female MZ pairs, aged 20–80 years, with blood lipid measurements available on both members were selected, and the sample consisted of pairs from Australia (459 pairs), Denmark (173 pairs), Finland (152 pairs), the Netherlands (331 pairs), Sweden (301 pairs), and the United Kingdom (462 pairs). DNA samples from one member of each pair were used for genotyping.
Verification of Zygosity
To verify the zygosity and monitor for possible sample mix-ups, a subset of the samples were subjected to genotyping of microsatellite or single nucleotide polymorphism (SNP) panels prior to the genome-wide analysis. The zygosity of all twin-pairs from the United Kingdom and Finland was verified by genotyping both twins of each twin-pair against a marker panel assaying of nine polymorphic autosomal (AC00818, D1S425, D2S338, D10S1651, D13S263, D16S539, D18S1102, D20S448, and GDB371492) and one X-linked microsatellite loci (DXS1227) in addition to the amelogenin locus yielding X- and Y-chromosomal-specific bands. The zygosity of the Danish samples was primarily assessed by nine DNA-based microsatellite markers with the PE Applied Biosystems AmpFISTR Profiler Plus Kit. For 10 twin-pairs, the zygosity was verified using the same microsatellite panel as described above. The zygosity of the Australian twin-pairs was verified using a commercial kit (AmpF1STR Profiler Plus Amplification KIT, Applied Biosystems, Foster City, CA) that analyzed nine independent, highly polymorphic DNA markers, plus the amelogenin marker for sex (Nyholt, Reference Nyholt2006). The zygosity of the Swedish twin-pairs was verified by genotyping of a validated panel of 47 SNPs as described in Hannelius et al. (Reference Hannelius, Gherman, Mäkelä, Lindstedt, Zucchelli, Lagerberg, Tybring, Kere and Lindgren2007).
Genotyping and Quality Control
The samples were genotyped using the Infinium II assay (Steemers et al., Reference Steemers, Chang, Lee, Barker, Shen and Gunderson2006) on the HumanHap300-Duo Genotyping BeadChips (Illumina Inc, San Diego, USA). Genotyping was performed according to the manufacturers’ instructions (Infinium II Assay Two-Sample Manual, #11230506 rev. A, Illumina Inc.). In total, 318,237 SNPs were genotyped on the BeadChips. The signal intensity data were converted into genotypes using the Illumina Beadstudio 2.0 software. Genotypes were assigned using custom-defined cluster positions generated from samples genotyped in-house. A total of 1,922 subjects were genotyped, of which two-thirds were processed and genotyped at the Finnish Genome Center in Helsinki and one-third at the SNP Technology Platform, Uppsala University, Uppsala, Sweden (www.genotyping.se). To verify the consistency of genotyping between laboratories, 14 duplicate samples were genotyped at both sites. The generated ~4.4 million genotypes were compared for consistency, yielding a reproducibility rate of 99.99%. The quality of samples and reagents was monitored by sample call rates, sample heterozygosity rates, and sex. Twenty-eight samples yielding <95% SNP call rates were discarded. Altogether, successful genotyping was performed on 1,868 unrelated subjects, of which 93.5% had a call rate of >99%. The average sample heterozygosity rate varied between 30.9% and 39.1%.
Imputation
Before imputation, individuals of non-European origin and with genotyping frequency of lower than 95% were excluded. These resulted in 1,720 individuals (Australia = 421, Denmark = 168, Finland = 137, the Netherlands = 275, Sweden = 297, and the United Kingdom = 422) in the final analysis set. SNPs having minor allele frequency of lower than 1%, Hardy Weinberg Equillibrium Fisher's exact test P value of lower than 1 × 10−6, or call rate lower than 99% were removed. The imputation was done using IMPUTE v2.0 (Howie et al., Reference Howie, Donnelly and Marchini2009) with reference haplotypes of HapMap 2 release 22 (http://hapmap.ncbi.nlm.nih.gov/downloads/phasing/2007-08_rel22/phased/).
Lipid Measurements
In all twin cohorts, we measured HDL, LDL, TC, and TG according to standard enzymatic methods. For a subset of twin-pairs, we also measured apolipoproteins A-I, A-II, B, and E, using immunoassays as described in Beekman et al. (Reference Beekman, Heijmans, Martin, Pedersen, Whitfield, DeFaire, van Baal, Snieder, Vogler, Slagboom and Boomsma2002).
Statistical Analysis
Because of a skewed distribution, TG and apolipoprotein A1, A2, B, and E values were log-transformed and, for all of phenotypes, outliers deviating more than three times the standard deviation from the trait mean were removed. Then means and absolute differences in lipid and apolipoprotein values were calculated for each pair. Both intra-pair differences and means were transformed into inverse normal distribution by rank for further analysis. Trait values were then adjusted for age (and differences additionally by mean effect) using a linear regression model. Association analyses between the imputed SNP probabilities and adjusted differences and means were conducted using SNPtest (Marchini et al., Reference Marchini, Howie, Myers, McVean and Donnelly2007). Cohort-specific results were then combined using the inverse variance meta-analysis method function in MetABEL (http://mga.bionet.nsc.ru/~yurii/ABEL/) package for statistical analysis program, R (http://cran.r-project.org/). We applied minor allele frequency threshold of 5% and imputation quality threshold of proper info >0.4 before meta-analysis.
Replication Samples
The Finnish replication twin set is a part of the Nicotine Addiction Genetics – Finland (FTC/NAG-FIN) study. FTC/NAG-FIN sample was ascertained from the Finnish Twin Cohort study consisting of adult twins born between 1938 and 1957 (www.twinstudy.helsinki.fi). Based on earlier health questionnaires, the twin-pairs concordant for ever-smoking were identified and recruited along with their family members (mainly siblings) for the NAG-FIN study (N = 2,265) as part of the consortium, including Finland, Australia, and United States (Broms et al., Reference Broms, Wedenoja, Largeau, Korhonen, Pitkäniemi, Keskitalo-Vuokko, Häppölä, Heikkilä, Heikkilä, Ripatti, Sarin, Salminen, Paunio, Pergadia, Madden, Kaprio and Loukola2012; Loukola et al., Reference Loukola, Broms, Maunu, Widén, Heikkilä, Siivola, Salo, Pergadia, Nyman, Sammalisto, Perola, Agrawal, Heath, Martin, Madden, Peltonen and Kaprio2008). Data collection took place between 2001 and 2005. The dataset was genotyped with Illumina human 670K chip in Wellcome Trust Sanger Institute.
The additional Swedish twins were from the TwinGene project, which is a part of the Swedish Twin registry. Twins born before 1958 were contacted to participate in a simple health check-up, with measurement of height, weight, waist and hip circumference, and blood pressure. Health and medication data were collected from self-reported questionnaires, and blood sampling materials were mailed to the subjects who then went to a local healthcare center for blood sampling for subsequent DNA extraction, serum collection, and clinical chemistry tests. Dataset was genotyped at the SNP Technology Platform in Uppsala University with Illumina OmniExpress 700K chip.
Results
The cohort-specific characteristics for the MZ twin sets are presented in Supplementary Table S1 online. The average age varies among the twin cohorts, being lowest in the Netherlands (mean age 33.6 years) and highest in Sweden (mean age 71.8 years). The MZ within-pair correlations in our dataset for lipids and lipoproteins are presented in Table 1. The within-pair correlations vary between the cohorts by their mean age as can be seen, for example, for the total cholesterol measures: For the oldest cohorts, Finnish and Swedish, the intra-pair correlations are 0.3 and 0.4, while for the younger cohorts the correlation is around 0.6–0.7 (Table 1). For this, as well as for the known age-trend in lipid levels, all the statistical analyses presented below are controlled for age.
TABLE 1 Within-Pair Correlations
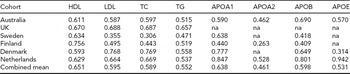
The within MZ twin-pair correlations for high-density lipoprotein (HDL) cholesterol, low-density lipoprotein (LDL) cholesterol, total cholesterol (TC), logarithmized triglyceride (TG), logarithmized apolipoproteins A1 (APOA1), A2 (APOA2), B (APOB), and E (APOE).
No adjustment for age was applied.
The combined means have been calculated using weights based on the size of the cohort.
na: not available.
We used the imputed genome-wide scan to identify genes associated with variability of lipids and apolipoproteins (see FIGURE 1). One locus having two SNPs with genome-wide significant effect (P < 5 × 10−8) on HDL variability was found (Table 2, FIGURE 2) and the effects were consistent across our six twin samples and in a joint analysis (Table 3). The variant with stronger association is located in an intron of the SRGAP2 gene on chromosome 1.
TABLE 2 Intra-Pair Difference Association Meta-Analysis Results

SNPs having P value < 1×10−6 in the association analysis of inverse normal transformed absolute intra pair differences.
SE: standard error of effect estimate, beta; N: number of individuals; HDL: high-density lipoprotein; TC: total cholesterol; APOA2: apolipoprotein A2.
TABLE 3 Effects of the Genome-Wide Significant SNPs in SRGAP2 Gene in the Six Twin Cohorts

Cohort wise results of the SNPs having P value < 5×10−7 in the association analysis of inverse normal transformed absolute intra pair differences. SE: standard error of the effect estimate, beta.

FIGURE 1 Quantile–quantile plot of all eight variability association analyses. Deviation from expected could be seen in HDL–cholesterol only.
QQ-PLOTS OF VARIABILITY ASSOCIATION ANALYSES

FIGURE 2 The upper panel shows –log10 (p-values) where the black dots are imputed and gray dots genotyped SNPs. In the second panel, the black line is the recombination rate (cM/Mb) and the red line is the distance from the best SNP measured as cM. The recombination rate and genetic positions are from http://hapmap.ncbi.nlm.nih.gov/downloads/recombination/2008-03_rel22_B36/rates/.
REGIONAL ASSOCIATION PLOT OF THE SRGAP2 LOCUS
We tried to replicate this association in two additional MZ twin datasets that showed positive effect for the C allele as in the initial analysis (Table 4), but with 50% smaller effect size. When combining results from the initial and replication analyses, the P value did not reach genome-wide significance (combined P value = 8.7 × 10−7). In addition to twin datasets, we followed the finding in a population cohort of 15,874 non-twin individuals with Bartlett's test of homogeneity of variance, but could not find support for the finding (combined P = 0.56, data not shown).
TABLE 4 Replication of rs2483058 Association on HDL-Variability in Additional Twin Datasets

HDL-variability association results for rs2483058 in two additional monozygotic twin datasets. SE: standard error of effect estimate, beta. N: number of individuals.
In addition to the one genome-wide significant hit, there was one suggestive hit (P = 8.16 × 10−7) for TC variability, rs17826288, which is positioned 66 kb downstream of the CD47 gene. Follow up of this locus in the population-based cohort resulted in a combined Bartlett's test P-value of 0.005, but the variances increased with the G allele, while in the MZGWA sample the variability decreases.
In addition, the aim of this study was to identify possible genes associated with apolipoprotein differences. No genome-wide significant hits were found, but we did find one suggestive hit associated with APOA2 intra-pair difference with a P value of 5.82 × 10−7. This SNP, rs6032680, is 20 kb downstream of CDH22 gene (Table 2). We could not replicate this finding, as the additional twin datasets and population-based cohort did not have measurement of apolipoproteins.
Results from the apolipoprotein level association screening are presented in Table 5. For the APOE/APOC1 locus, there was one SNP that had a genome-wide significant association for APOB and APOE. In addition, we found one new genome-wide significant locus for apolipoprotein levels, DOCK1, which had an association with APOA2 levels. There was also suggestive deviation from the expected P values (supplementary Figure S1 online) for APOA1, but no genome-wide significant loci were found.
TABLE 5 Genome-Wide Significant Association Analysis Results on Apolipoprotein Levels

SNPs having P value of lower than 5×10−8 in the association analysis of intra pair means of apolipoproteins. N: number of individuals; SE: standard error of the effect estimate, beta; APOA2: apolipoprotein A2; APOB: apolipoprotein B; APOE = apolipoprotein E.
Discussion
It has been shown previously (Visscher & Posthuma, Reference Visscher and Posthuma2010) that the MZ difference approach has more power than the population variance method when the within-pair correlation of twins is higher than 0.3, which is the case for both circulating blood lipids and apolipoproteins (Table 1). Following the formulae in the study by Visscher and Posthuma (Reference Willer, Sanna, Jackson, Scuteri, Bonnycastle, Clarke, Heath, Timpson, Najjar, Stringham, Strait, Duren, Maschio, Busonero, Mulas, Albai, Swift, Morken, Narisu, Bennett, Parish, Shen, Galan, Meneton, Hercberg, Zelenika, Chen, Li, Scott, Scheet, Sundvall, Watanabe, Nagaraja, Ebrahim, Lawlor, Ben-Shlomo, Davey-Smith, Shuldiner, Collins, Bergman, Uda, Tuomilehto, Cao, Collins, Lakatta, Lathrop, Boehnke, Schlessinger, Mohlke and Abecasis2010), with a within-pair correlation of 0.651 (0.605 for CC homozygous pairs), which was derived from the MZ dataset, we get a multiplicative effect of 1.17 in population variances. For a hit with a multiplicative effect of 1.2 in variances, the relative efficiency is approximately two to three times higher for the twin approach and the number of MZ pairs used in this (1,720) study corresponds to a population sample of 11,000.
Large international efforts combining results from multiple cohorts can achieve the power to detect variability of G × E associations by increasing the number of individuals in the analysis, but at the same time, more variability is induced by population-specific effects. One of the challenges in gene–environment interaction mapping with quantitative traits is the amount of extra variability in the analysis because of the measurement error of the environmental factor. However, when mapping variability genes instead of G × E interactions, this is not an issue because no specific environmental factor is assumed.
We can infer from twin data that the differential variability is caused by differential sensitivity to the environment or lifestyle and not by other interacting genes, since the MZ twins are essentially genetically identical at the sequence level. With our genome-wide MZ twin dataset we showed initial evidence for a locus associated with twins being different in their HDL values depending on their genotypes in SRGAP2 gene. The SRGAP2 has been connected to multiple different cancers, but there is no known connection between the gene and lipid metabolism.
Because of the restricted availability of MZ twin datasets, we additionally tried to replicate the genome-wide significant finding in population based cohorts but could not find support. This failed replication could be due to false positive finding or power issues in homogeneity of variance analysis with Bartlett's test. The suggestive TC variability locus, CD47, showed some evidence of variability association in the population cohorts, but in opposite direction.
By the analysis of MZ-twin apolipoprotein levels, we were able to replicate one previously published lipid gene, but also found a new locus associated with APOA2. As this is one of the first GWA analyses studying apolipoprotein fractions in addition to lipids, further replication studies for the new locus, DOCK1, are called for.
The mechanisms of joint actions of the gene and lifestyle factors are unknown, but these suggestive results provide basis for further functional and epidemiological studies. Further, detailed consideration of environmental factors affecting lipid levels as well as developmental changes over the lifespan need to be incorporated in the analysis models.
Author Contributions
MP, A-CS, AP, JK, KOK, NLP, DIB, TS, NGM, SR, and LP conceived and designed the experiments. JBW, MP, PMV, GWM, MF, GW, EJCdG, PKEM, KC, TIAS, KHP, TR, KS, EW, UJ, A-CS, and AP performed the experiments. IS, MP, JM, IR, and SR analyzed the data. SR, MP, IS, NGM, PMV, GWM, MF, PKEM, KS, EW, UL, A-CS, AP, DIB, and LP contributed reagents/materials/analysis tools. All authors wrote and approved the final version of the paper.
Supplementary Material is available on the Cambridge Journals Online website.
Acknowledgments
The GenomEUtwin project is supported by the European Commission under the program “Quality of Life and Management of the Living Resources” of 5th Framework Programme (QLG2-CT-2002–01254).
The Swedish Twin Registry was supported by the US National Institutes of Health (AG028555, AG08724, AG04563, AG10175, AG08861), the Swedish Research Council, the Swedish Heart-Lung Foundation, the Swedish Foundation for Strategic Research, the Royal Swedish Academy of Science, and ENGAGE (within the European Union Seventh Framework Programme, HEALTH – F4-2007-201413).
The Danish Twin Registry has been supported by the Danish Medical Research Council, the Danish Diabetes Foundation, the Danish Heart Association, and the Novo Nordic Foundation.
The TWINSUK study was funded by the Wellcome Trust (Grant ref. 079771); European Community's Framework 6 Project EUroClot. The study also received support from the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St Thomas’ NHS Foundation Trust in partnership with King's College London. Timothy D Spector is an NIHR senior investigator and is a holder of an ERC Advanced Principal Investigator award.
The Australian Twin Cohort was funded by the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, 552498), the Australian Research Council (A7960034, A79906588, A79801419, DP0770096, DP0212016, DP0343921), and the US National Institutes of Health (AA07535, AA10248, AA11998, AA13320, AA13321, AA13326, AA14041, AA17688, DA12854, MH66206).
The Netherlands Twin Registry (NTR) has been supported by the Netherlands Scientific Organization (911–09–032, 480–04–004, CMSB NOW Genomics, NWO–SPI 56–464–1419, NBIC/BioAssist/RK/2008.024), BBMRI–6 NL, Institute for Health and Care Research (EMGO+), the European Union (EU/WLRT–2001–01254, ERC Advanced 230374).
The Finnish Twin Cohort Study was supported by the Academy of Finland Center of Excellence in Complex Disease Genetics (grant numbers 213506, 129680), the Finnish Foundation for Cardiovascular Research, the Finnish Academy (grant numbers 129494, 139635, and 251217), and the Sigrid Juselius Foundation.
Platform in Uppsala is funded by Uppsala University and Uppsala University Hospital and the Swedish Wallenberg Foundation.
SR was supported by the Academy of Finland Center of Excellence in Complex Disease Genetics (213506 and 129680), Academy of Finland (251217), the Finnish foundation for Cardiovascular Research, and the Sigrid Juselius Foundation. MP is partly financially supported for this work by the Finnish Academy SALVE program “Pubgensense” 129322 and by grants from Finnish Foundation for Cardiovascular Research. KP was funded by Helsinki University Central Hospital grants. TR has been funded from the Academy of Finland Research Programme on Ageing, Finnish Ministry of Education.
We thank the staff of the SNP Platform for their contributions to genotyping.









