Introduction
Substance use stands as a major risk factor contributing significantly to the global burden of disease (Degenhardt et al., Reference Degenhardt, Charlson, Ferrari, Santomauro, Erskine, Mantilla-Herrara and Vos2018). To improve prevention and treatment, it is important to increase our understanding of the pathogenesis of substance use behaviors (SUBs) (e.g. initiation, lifetime use, and severity of consumption) and substance use disorders (SUDs). This includes identifying factors influencing different aspects of substance use, from severity of substance consumption to pathological substance use.
Evidence from developmental psychology suggests two main pathways for substance use initiation and escalation towards SUDs: an externalizing pathway characterized by high levels of impulsivity, aggression, and sensation-seeking, and an internalizing pathway characterized by using substances to cope with negative affect, depression, and anxiety (Hussong, Jones, Stein, Baucom, & Boeding, Reference Hussong, Jones, Stein, Baucom and Boeding2011; Krueger et al., Reference Krueger, Hicks, Patrick, Carlson, Iacono and McGue2002). This is supported by prospective evidence of distinct externalizing and internalizing pathways leading to future SUBs and SUDs (Meque, Dachew, Maravilla, Salom, & Alati, Reference Meque, Dachew, Maravilla, Salom and Alati2019; Ning, Gondek, Patalay, & Ploubidis, Reference Ning, Gondek, Patalay and Ploubidis2020). However, evidence is weaker for the internalizing pathway. This may be due to comorbidity between internalizing and externalizing problems and differences regarding gender, developmental timing, or type of substance use (Heitzeg, Hardee, & Beltz, Reference Heitzeg, Hardee and Beltz2018; Ning et al., Reference Ning, Gondek, Patalay and Ploubidis2020). For instance, internalizing traits seem to be related to reduced alcohol consumption in middle adulthood subjects, but to increased consumption in more severe outcomes (i.e. heavy drinking and alcohol use disorder [AUD]) during transition to adulthood (Ning et al., Reference Ning, Gondek, Patalay and Ploubidis2020).
Additional support for a causal effect of both externalizing and internalizing problems on SUBs and SUDs comes from the Mendelian randomization studies. Mendelian randomization leverages information from molecular genetic studies to infer causal relationships between phenotypes (Chen, Tubbs, Liu, Thach, & Sham, Reference Chen, Tubbs, Liu, Thach and Sham2024). This approach found evidence for a causal effect of attention-deficit/hyperactivity disorder (ADHD) on tobacco and cannabis use behaviors, but also for reverse causation (Treur et al., Reference Treur, Demontis, Smith, Sallis, Richardson, Wiers and Munafò2019; Vilar-Ribó et al., Reference Vilar-Ribó, Sánchez-Mora, Rovira, Richarte, Corrales, Fadeuilhe and Soler Artigas2020). It also indicated that depression and neuroticism are risk factors for tobacco use and alcohol dependence and discarded reverse causation (Polimanti et al., Reference Polimanti, Peterson, Ong, MacGregor, Edwards, Clarke and Derks2019; Sallis, Davey Smith, & Munafò, Reference Sallis, Davey Smith and Munafò2019; Yao et al., Reference Yao, Xu, Cai, Liu, Ma, Li and Li2021). However, there is also some evidence for a bidirectional causal relationship between depression and smoking behavior (Wootton et al., Reference Wootton, Richmond, Stuijfzand, Lawn, Sallis, Taylor and Munafò2020).
Twin and family studies have estimated heritability of around 40–60% for different SUDs, partly specific and partly shared between substances (Deak & Johnson, Reference Deak and Johnson2021). Genome-wide association studies (GWAS) revealed that a considerable portion of the heritability is due to the additive effect of many common variants of low individual effect, that is SUDs are highly polygenic traits. Many of these genetic variants are shared between SUDs and among SUDs and related phenotypes (Deak & Johnson, Reference Deak and Johnson2021; Gelernter & Polimanti, Reference Gelernter and Polimanti2021). Polygenic scores (PGS) may be used to estimate the risk of SUD as the sum of susceptibility alleles carried by an individual weighted by their effect. Cross-trait PGS confirmed the existence of shared genetic susceptibility between related phenotypes. For instance, PGS of the average number of alcoholic drinks consumed per week (DrnkWk), major depressive disorder (MDD), and ADHD are associated with AUD (Facal et al., Reference Facal, Flórez, Blanco, Rodríguez, Pereiro and Fernández2021), PGS of ADHD is associated with tobacco use (Leppert et al., Reference Leppert, Millard, Riglin, Davey Smith, Thapar, Tilling and Stergiakouli2020), and PGS of smoking initiation with DrnKWk, nicotine dependence and conduct disorder (Chang et al., Reference Chang, Whitfield, Liu, Medland, Hickie, Martin and Gillespie2019).
Among the different methods to measure shared genetic susceptibility, cross-trait Linkage Disequilibrium (LD) score regression has reached enormous popularity in recent years due to the main advantage that it relies only on GWAS summary statistics. LD score regression calculates the correlation coefficient of additive genetic effects for two traits across the genome (genetic correlation, rg) assuming a simple infinitesimal model, that is all SNPs are causal with normally distributed additive genetic effects (Bulik-Sullivan et al., Reference Bulik-Sullivan, Finucane, Anttila, Gusev, Day, Loh and Neale2015). Recent studies have examined the genetic correlation between different SUBs and SUDs using LD score regression. They have shown that smoking severity, defined as the number of cigarettes smoked per day (Cigsperday), and nicotine dependence are almost genetically identical (rg = 0.95) (Quach et al., Reference Quach, Bray, Gaddis, Liu, Palviainen, Minica and Hancock2020). In fact, Cigsperday was proposed as a ‘genetic proxy’ for nicotine dependence, considering their strong genetic correlation and their similar pattern of associations with multiple diseases (Sanchez-Roige, Cox, Johnson, Hancock, & Davis, Reference Sanchez-Roige, Cox, Johnson, Hancock and Davis2021). By contrast, the genetic correlation between alcohol consumption, measured as the number of drinks per week (DrnkWk), and alcohol dependence was weaker (rg = 0.37) (Walters et al., Reference Walters, Polimanti, Johnson, McClintick, Adams, Adkins and Agrawal2018). Similarly, there is a moderate genetic correlation between smoking initiation and nicotine dependence (rg = 0.40) (Quach et al., Reference Quach, Bray, Gaddis, Liu, Palviainen, Minica and Hancock2020). Low to moderate genetic correlation also exists between SUBs involving different substances, such as smoking initiation and DrnkWk (rg = 0.27) (Saunders et al., Reference Saunders, Wang, Chen, Jang, Liu, Wang and Vrieze2022).
Significant genetic correlations have also been identified between both externalizing and internalizing traits and both SUBs and SUDs across different substances. For example, ADHD was genetically correlated with SUDs such as nicotine dependence, alcohol dependence and cannabis use disorder (CUD) (Abdellaoui, Smit, van den Brink, Denys, & Verweij, Reference Abdellaoui, Smit, van den Brink, Denys and Verweij2021; Vink, Treur, Pasman, & Schellekens, Reference Vink, Treur, Pasman and Schellekens2020; Walters et al., Reference Walters, Polimanti, Johnson, McClintick, Adams, Adkins and Agrawal2018); as well as with SUBs such as tobacco and cannabis use (Liu et al., Reference Liu, Jiang, Wedow, Li, Brazel, Chen and Vrieze2019; Pasman et al., Reference Pasman, Verweij, Gerring, Stringer, Sanchez-Roige, Treur and Vink2018; Soler Artigas et al., Reference Soler Artigas, Sánchez-Mora, Rovira, Richarte, Garcia-Martínez, Pagerols and Ribasés2020). Other risky-behavior traits considered as externalizing were associated with SUBs. For example, age at first sexual intercourse (AFS) and age at first birth (AFB) showed a significant genetic correlation with age of smoking initiation and cannabis use (Mills et al., Reference Mills, Tropf, Brazel, van Zuydam, Vaez, Agbessi and Day2021). Further, other studies reported a significant genetic correlation of depression with both SUBs and SUDs, including smoking initiation, smoking cessation, nicotine dependence, and alcohol dependence (Abdellaoui et al., Reference Abdellaoui, Smit, van den Brink, Denys and Verweij2021; Liu et al., Reference Liu, Jiang, Wedow, Li, Brazel, Chen and Vrieze2019; Vink et al., Reference Vink, Treur, Pasman and Schellekens2020; Walters et al., Reference Walters, Polimanti, Johnson, McClintick, Adams, Adkins and Agrawal2018).
Despite the insight provided by genome-wide genetic correlation to understand shared genetic susceptibility among related phenotypes, it has a clear limitation. Genome-wide genetic correlation, as estimated by LD score regression, can capture genetic overlap between two phenotypes only when the majority of shared variants exhibit a strong net correlation in the direction of effects relative to each other (i.e. effects are correlated in the same or opposite directions in the two phenotypes). However, it does not allow for the assessment of genetic overlap if the shared variants reveal a balanced mixture of concordant and discordant directions of effects. The bivariate causal mixture model (MiXeR) overcomes this limitation, characterizing the pattern of genetic overlap between two traits beyond genome-wide genetic correlation (Frei et al., Reference Frei, Holland, Smeland, Shadrin, Fan, Maeland and Dale2019). First, MiXeR estimates the number of causal variants involved in each trait, instead of assuming an infinitesimal model. Next, MiXeR estimates the number of shared variants between the two traits and the genetic correlation of shared variants. Therefore, in contrast to genome-wide genetic correlation studies, MiXeR enables the discovery of genetic overlap even if the shared variants possess a mixture of concordant and discordant effect directions.
In the current study, we aimed to characterize the genetic relationship of SUBs and SUDs with externalizing and internalizing traits using MiXeR. Specifically, we investigated if: (i) SUBs and SUDs differ in their pattern of polygenic overlap with externalizing and internalizing traits, including the proportion of genetic overlap and genetic correlation of shared variants, and (ii) these differences are present regardless of the particular substance involved. This approach can improve our understanding of the contribution of the externalizing and internalizing pathways to SUBs and SUDs.
Methods
Genome-wide association studies (GWAS)
We included summary statistics for SUBs and SUDs and externalizing and internalizing traits from large GWAS (Table 1 and Supplementary Table 1). These GWAS were selected according to their large sample size and public availability. All GWAS summary statistics were based on samples of European ancestry to share similar linkage disequilibrium patterns, a requirement for MiXeR analysis.
Table 1. GWAS summary statistics used in MiXeR analysis

For the three most studied drugs, tobacco, alcohol, and cannabis, we selected at least one SUB and one SUD or SUD-proxy. As tobacco use behavior, we chose ever be a regular smoker (EverSmk) (Saunders et al., Reference Saunders, Wang, Chen, Jang, Liu, Wang and Vrieze2022), which reflects smoking initiation. To assess tobacco use disorder (TUD), we generated our own GWAS of TUD using UK Biobank data (further information in Supplementary Methods). UK Biobank data was obtained under accession number 27412. We also used Cigsperday (Saunders et al., Reference Saunders, Wang, Chen, Jang, Liu, Wang and Vrieze2022), which is a good ‘genetic proxy’ of nicotine dependence (Sanchez-Roige et al., Reference Sanchez-Roige, Cox, Johnson, Hancock and Davis2021). As alcohol use behaviors, we selected DrnkWk (Saunders et al., Reference Saunders, Wang, Chen, Jang, Liu, Wang and Vrieze2022), a score comprising the items related to alcohol consumption of the Alcohol Use Disorders Identification Test (AUDIT-C) (Sanchez-Roige et al., Reference Sanchez-Roige, Palmer, Fontanillas, Elson, Adams, Howard and Wilson2019) and AUDIT-P (Sanchez-Roige et al., Reference Sanchez-Roige, Palmer, Fontanillas, Elson, Adams, Howard and Wilson2019). AUD was assessed by an AUD GWAS (Icick et al., Reference Icick, Shadrin, Holen, Karadag, Parker, O’Connell and Andreassen2023). For cannabis, we selected lifetime cannabis use (CANN) (Pasman et al., Reference Pasman, Verweij, Gerring, Stringer, Sanchez-Roige, Treur and Vink2018) and CUD (Johnson et al., Reference Johnson, Demontis, Thorgeirsson, Walters, Polimanti, Hatoum and Børglum2020).
As externalizing traits, we chose those four traits that were previously used to construct an externalizing genetic factor and do not involve substance use (Karlsson Linnér et al., Reference Karlsson Linnér, Mallard, Barr, Sanchez-Roige, Madole, Driver and Dick2021): ADHD (Demontis et al., Reference Demontis, Walters, Athanasiadis, Walters, Therrien, Nielsen and Børglum2023), AFS (Mills et al., Reference Mills, Tropf, Brazel, van Zuydam, Vaez, Agbessi and Day2021), number of sexual partners (NSEX) (Karlsson Linnér et al., Reference Karlsson Linnér, Biroli, Kong, Meddens, Wedow, Fontana and Wagner2019), and general risk tolerance (RISK) (Karlsson Linnér et al., Reference Karlsson Linnér, Biroli, Kong, Meddens, Wedow, Fontana and Wagner2019). In addition, AFB (Mills et al., Reference Mills, Tropf, Brazel, van Zuydam, Vaez, Agbessi and Day2021) and childhood aggressive behavior (AGG) (Ip et al., Reference Ip, van der Laan, Krapohl, Brikell, Sánchez-Mora, Nolte and Boomsma2021), related to externalizing behavior (Baselmans et al., Reference Baselmans, Hammerschlag, Noordijk, Ip, van der Zee, de Geus and van ‘t Ent2021; Mills et al., Reference Mills, Tropf, Brazel, van Zuydam, Vaez, Agbessi and Day2021), were also included.
For internalizing traits, different phenotypes of depression and anxiety were selected: a broader diagnosis of depression (DEP) (Howard et al., Reference Howard, Adams, Clarke, Hafferty, Gibson, Shirali and McIntosh2019), MDD (Wray et al., Reference Wray, Ripke, Mattheisen, Trzaskowski, Byrne, Abdellaoui and Sullivan2018), a continuous trait for anxiety based on the Generalized Anxiety Disorder 2-item scale (GAD) (Levey et al., Reference Levey, Gelernter, Polimanti, Zhou, Cheng, Aslan and Stein2020) and self-reported anxiety (ANX) (http://www.nealelab.is/uk-biobank/). Moreover, neuroticism, a core feature of internalizing behavior (Griffith et al., Reference Griffith, Zinbarg, Craske, Mineka, Rose, Waters and Sutton2010), measured by a neuroticism score based on 12 items (NEURO) was also included (Nagel, Jansen, et al., Reference Nagel, Jansen, Stringer, Watanabe, De Leeuw, Bryois and Posthuma2018). Specific subclusters and items of NEURO were also analyzed: depressed affect subcluster of NEURO (DEP-NEURO) (Nagel, Jansen, et al., Reference Nagel, Jansen, Stringer, Watanabe, De Leeuw, Bryois and Posthuma2018), worry subcluster of NEURO (WORRY-NEURO) (Nagel, Jansen, et al., Reference Nagel, Jansen, Stringer, Watanabe, De Leeuw, Bryois and Posthuma2018), mood instability (MOOD) (Nagel, Watanabe, Stringer, Posthuma, & Van Der Sluis, Reference Nagel, Watanabe, Stringer, Posthuma and Van Der Sluis2018), and often feeling lonely (LONE) (Nagel, Watanabe, et al., Reference Nagel, Watanabe, Stringer, Posthuma and Van Der Sluis2018). The two subclusters of NEURO show distinct patterns of genetic correlation with behavioral, neuropsychiatric, anthropometric, and health-related phenotypes, while a genetic correlation of NEURO is a mix of both subclusters (Nagel, Jansen, et al., Reference Nagel, Jansen, Stringer, Watanabe, De Leeuw, Bryois and Posthuma2018; Nagel, Watanabe, et al., Reference Nagel, Watanabe, Stringer, Posthuma and Van Der Sluis2018). Subjective well-being (SWB) (Kim et al., Reference Kim, Kim, Hwang, Ko, Jung, Shim and Won2022) was also selected as it is highly correlated with anxious/depressive behavior (Bartels, Cacioppo, Van Beijsterveldt, & Boomsma, Reference Bartels, Cacioppo, Van Beijsterveldt and Boomsma2013). Other aspects of loneliness were also included: the frequency of being able to confide in someone close (CONF) (http://www.nealelab.is/uk-biobank/) and a combined multi-trait GWAS of loneliness and social isolation (LONE-MTAG) (Day, Ong, & Perry, Reference Day, Ong and Perry2018), which is strongly genetically correlated with neuroticism and depressive symptoms.
Data analysis
We applied MiXeR, version 1.2.0, to characterize the pattern of genetic overlap between SUBs and SUDs and externalizing and internalizing traits using GWAS summary statistics (Frei et al., Reference Frei, Holland, Smeland, Shadrin, Fan, Maeland and Dale2019). First, univariate MiXeR was applied to each trait to estimate their polygenicity, that is the fraction of causal variants in the genome, and discoverability, that is the causal effect size variance, maximizing likelihood estimation (Holland et al., Reference Holland, Frei, Desikan, Fan, Shadrin, Smeland and Dale2020). SNP-based heritability (h2SNP) is derived from the product of these estimates, taking heterozygosity into account (Frei et al., Reference Frei, Holland, Smeland, Shadrin, Fan, Maeland and Dale2019). Polygenicity is presented as the number of causal variants required to explain 90% of h2SNP. This threshold of 90% is applied to exclude those variants with infinitesimally small effects. Next, bivariate MiXeR was applied to assess the pattern of genetic overlap between each pair of traits. Using the parameters estimated in univariate MiXeR for each trait, bivariate MiXeR estimates the number of shared causal variants, the number of unique causal variants specific to each trait and the correlation of effect sizes of shared variants (rgs). To enhance result interpretation, we assessed the degree of genetic overlap between each pair of traits by quantifying the percentage of shared variants with respect to the SUB/SUD. To assess model fit, the difference between the Akaike information criterion (AIC) for the reference model and the MiXeR-fitted model was calculated. Positive AIC differences indicate that the MiXeR-fitted estimates are more informative than the reference model. The minimum and maximum possible overlap are used as the reference models in bivariate MiXeR. The minimum possible overlap between two traits is restricted by their genome-wide genetic correlation and the maximum possible overlap is restricted by the polygenicity of the least polygenic trait. Further information about MiXeR is provided in Supplementary Methods and the original publication (Frei et al., Reference Frei, Holland, Smeland, Shadrin, Fan, Maeland and Dale2019).
It is well known that results for methods based on summary statistics are dependent on the power of the GWAS. Underpowered GWAS may lead to unreliable results. To limit bivariate MiXeR analyses to the most well-powered GWAS, we selected GWAS according to their univariate MiXeR model fit. Specifically, we selected those traits with substantial improvement in model fit by the causal mixture model in comparison to the infinitesimal model, which assumes all variants as causal. An arbitrary threshold of 50 as the difference in AIC between the infinitesimal model and the causal mixture model was applied. The difference in AIC between these two models is strongly correlated with the power of a GWAS, roughly estimated as the product of the effective sample size and h2SNP in our data (Pearson’s correlation coefficient = 0.92, 95% CI: 0.83–0.96) (Supplementary Methods and Supplementary Figure 1). Using BIC as an alternative measure of model fit would lead to a similar selection (Supplementary Table 2). For Cigsperday GWAS, we excluded the cluster of neuronal nicotinic acetylcholine receptor (nAchR) genes at chromosome 15 because its strong effect on this phenotype violates MiXeR’s assumption that the effect sizes of causal variants are normally distributed. Further details about the exclusion of the cluster of nAchR genes at chromosome 15 for Cigsperday are shown in Supplementary Methods.
Results
After excluding GWAS with poor MiXeR model fit, we included the following tobacco and alcohol use behaviors, respectively: EverSmk and DrnkWk; and a SUD or SUD-proxy for each of these substances: Cigsperday and AUD (Figure 1). We included five externalizing traits: AFS, NSEX, AFB, RISK, and ADHD; and five internalizing traits: NEURO, DEP-NEURO, WORRY-NEURO, DEP, and MOOD.

Figure 1. GWAS filtering according to their univariate MiXeR AIC. GWAS selected for MiXeR analyses, that is, those with a difference between the AIC calculated for the infinitesimal model and the AIC calculated for the causal mixture model higher than 50, are highlighted in bold. The dashed line represents univariate MiXeR AIC difference = 50. Traits are described in Table 1 and Supplementary Table 1. ADHD, attention deficit hyperactivity disorder; AFB, age at first birth; AFS, age at first sexual intercourse; AGG, childhood aggressive behavior; ANX, self-response to ever being diagnosed by anxiety, nerves or generalized anxiety disorder; AUD, alcohol use disorder; AUDIT-C, items related to alcohol consumption of the alcohol use disorders identification test; AUDIT-P, items related to problematic consequences of drinking of the alcohol use disorders identification test; CANN, lifetime cannabis use; Cigsperday, average number of cigarettes smoked per day; CONF, frequency of being able to confide in someone close; CUD, cannabis use disorder; DEP, lifetime diagnosis of major depressive disorder or broad self-reported depression; DEP-NEURO, depressed affect subcluster of neuroticsm; DrnkWk, average number of drinks drunk each week; EverSmk, ever been a regular smoker; GAD, generalized anxiety disorder; LONE, often feeling lonely; LONE-MTAG, combined multi-trait GWAS of loneliness and social isolation; MDD, major derepssive disorder; MOOD, experiencing mood swings; NEURO, neuroticism; NSEX, lifetime number of sexual partners; RISK, general risk tolerance; SWB, subjectrive well-being; TUD, tobacco use disorder; WORRY-NEURO, worry subcluster of neuroticism.
Univariate MiXeR results
Univariate MiXeR estimated the polygenicity and discoverability of each of the selected traits (Supplementary Table 2). Among SUBs and SUDs, EverSmk was the most polygenic trait (N = 11.6 K causal variants explaining 90% of the h2SNP, SE = 0.7 K). DrnkWk and AUD had similar polygenicity (DrnkWk: N = 7.7 K, SE = 0.6 K; AUD: N = 7.7 K, SE = 0.3 K). Cigsperday was the least polygenic trait with 4.3 K (SE = 0.6 K) variants involved. All selected externalizing and internalizing traits were highly polygenic with 9.7 K–12.2 K variants explaining 90% of the h2SNP, except for ADHD with 7.7 K (SE = 0.4 K) causal variants.
Bivariate MiXeR results
Bivariate MiXeR was used to estimate the pattern of genetic overlap between each of the four selected SUBs and SUDs and the 10 selected externalizing and internalizing traits. Specifically, the percentage of shared variants with respect to the SUB/SUD and the genetic correlation of shared variants were estimated for each pair of traits. Full results can be found in Supplementary Table 3.
Bivariate MiXeR model fit
For almost all bivariate analyses, both AIC compared to minimum and maximum overlap were positive, indicating a good model fit (Supplementary Table 3). Minimum AIC was negative for AUD with NSEX and DEP, indicating that the overlap modeled by MiXeR was not distinguishable from the minimum overlap taking into account the genetic correlation between both traits. Maximum AIC was negative for some analyses for which there was a high overlap between both traits, indicating that the overlap estimated by MiXeR was not distinguishable from complete genetic overlap.
Pattern of genetic overlap between externalizing/internalizing traits and SUBs
The percentage of SUB causal variants shared with externalizing traits ranged from 52.0% to 95.9% (Figure 2 and Supplementary Figure 2). The genetic correlation of shared variants was moderate to high with almost all externalizing traits (absolute values of rgs = 0.28–0.87), except between DrnkWk and AFB (absolute value of rgs = 0.05) (Figure 3).
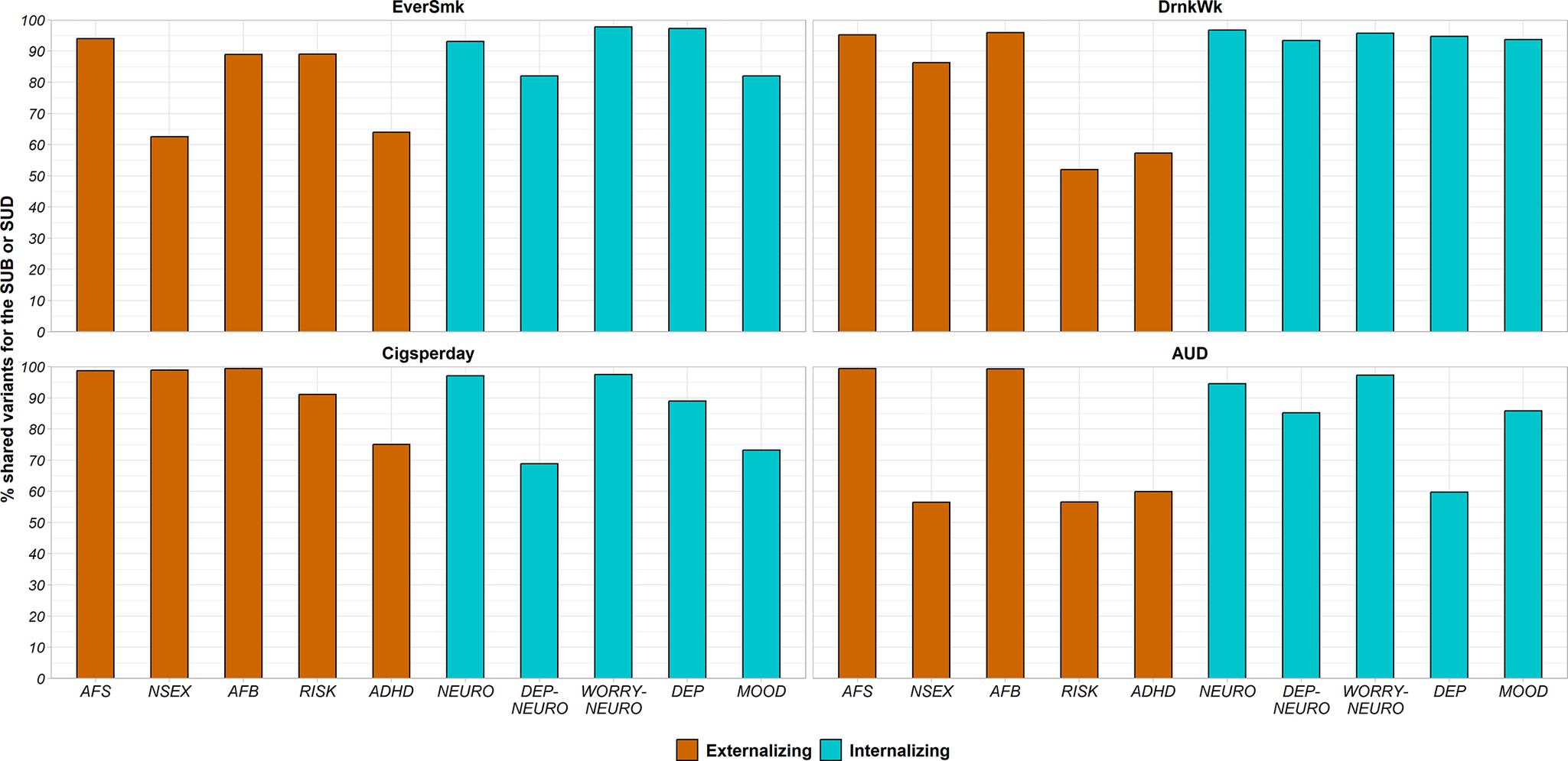
Figure 2. Percentage of variants of each SUB (EverSmk and DrnkWk) or SUD/SUD-proxy (Cigsperday and AUD) shared with each externalizing (colored in orange) or internalizing (colored in green) trait. Traits are described in Table 1. ADHD, attention deficit hyperactivity disorder; AFB, age at first birth; AFS, age at first sexual intercourse; AUD, alcohol use disorder; Cigsperday, average number of cigarettes smoked per day; DEP, lifetime diagnosis of major depressive disorder or broad self-reported depression; DEP-NEURO, depressed affect subcluster of neuroticism; DrnkWk, average number of drinks drunk each week; EverSmk, ever been a regular smoker; MOOD, experiencing mood swings; NEURO, neuroticism; NSEX, lifetime number of sexual partners; RISK, general risk tolerance; WORRY-NEURO, worry subcluster of neuroticism.

Figure 3. Genetic correlation of the shared variants (rgs) between each SUB (EverSmk and DrnkWk) or SUD/SUD-proxy (Cigsperday and AUD) and each externalizing (colored in orange) or internalizing (colored in green) trait. Traits are described in Table 1. Error bars represent ±1 SD. The direction was reversed for AFB and AFS to indicate the genetic correlation with a higher externalizing behavior. ADHD, attention deficit hyperactivity disorder; AFB, age at first birth; AFS, age at first sexual intercourse; AUD, alcohol use disorder; Cigsperday, average number of cigarettes smoked per day; DEP, lifetime diagnosis of major depressive disorder or broad self-reported depression; DEP-NEURO, depressed affect subcluster of neuroticism; DrnkWk, average number of drinks drunk each week; EverSmk, ever been a regular smoker; MOOD, experiencing mood swings; NEURO, neuroticism; NSEX, lifetime number of sexual partners; RISK, general risk tolerance; WORRY-NEURO, worry subcluster of neuroticism.
In the case of internalizing traits, genetic overlap with SUBs was high (Figure 2 and Supplementary Figure 2). EverSmk and DrnkWk shared more than 82% and 93% of their causal variants with internalizing traits, respectively (Figure 2). The genetic correlation of shared variants with internalizing traits was clearly lower than with externalizing traits (absolute values of rgs = 0.02–0.35) (Figure 3).
Despite following the same general pattern of genetic overlap, some differences emerged between tobacco use behavior (EverSmk) and alcohol use behavior (DrnkWk). EverSmk exhibited a slightly higher genetic correlation of shared variants (absolute values of rgs = 0.40–0.87) with externalizing traits compared to DrnkWk (absolute values of rgs = 0.05–0.81) (Figure 3). Similarly, internalizing traits exhibited a higher genetic correlation of shared variants with EverSmk (absolute values of rgs = 0.06–0.35) compared to DrnkWk (absolute values of rgs = 0.02–0.11). Notably, DrnkWk showed a negative genetic correlation of shared variants, although close to 0, with all internalizing traits except for DEP, while EverSmk only displayed a negative genetic correlation of shared variants with WORRY-NEURO among internalizing traits.
Pattern of genetic overlap between externalizing/internalizing traits and SUDs/SUD-proxies
The percentage of causal variants for SUDs and SUD-proxies shared with externalizing traits ranged from 56.5% to 99.4% (Figure 2 and Supplementary Figure 2). The genetic correlation of shared variants for SUDs and SUD-proxies with externalizing traits was relatively high (absolute values of rgs = 0.25–0.95) (Figure 3). In all pairs, a higher value for externalizing behavior was positively correlated with both SUBs and SUDs among the shared variants (Figure 3).
Genetic overlap with internalizing traits for SUDs and SUD-proxies (59.7%–97.4%) was more variable than for SUBs (82.0%–97.8%) (Figure 2 and Supplementary Figure 2). Cigsperday and AUD demonstrated a higher genetic correlation of shared variants with internalizing traits, except WORRY-NEURO, (absolute values of rgs = 0.23–0.98) than SUBs (absolute values of rgs = 0.02–0.35) (Figure 3). In contrast to the remaining internalizing traits, WORRY-NEURO showed a similar pattern of genetic overlap with both SUBs and SUDs. WORRY-NEURO shared a large number of variants (>95%) with all SUBs and SUDs, despite possessing low genetic correlation of shared variants (Figure 2 and Figure 3). The direction of the genetic correlation of shared variants was positive for pairs involving AUD and internalizing traits, contrary to DrnkWk (Figure 3). By contrast, the direction of the genetic correlation of shared variants with internalizing traits was positive for both tobacco traits.
Relationship between the genetic correlation of shared variants and the number of shared causal variants
An inverse relationship was observed between the genetic correlation of shared variants and the number of shared causal variants for each pair of traits, with a significant negative correlation between these variables (r = −0.66, P = 4.32x10−6) (Figure 4). Those pairs of traits exhibiting a lower genetic correlation of shared variants than expected given the number of shared variants comprised predominantly SUBs and internalizing traits (small open triangles and squares in Figure 4). In contrast, pairs showing a genetic correlation of shared variants higher than expected based on the number of shared variants involved SUDs or SUD-proxies with externalizing traits (large filled triangles and squares in Figure 4).

Figure 4. Scatter plot illustrating the negative relationship between the genetic correlation of shared variants (rgs) and the percentage of shared variants across analyzed trait pairs. The solid line represents the linear regression, while the shaded region depicts the 95% confidence interval. On the x-axis, the percentage of shared variants between each SUB or SUD and each externalizing or internalizing trait with respect to the SUB/SUD is displayed. The y-axis shows the genetic correlation of shared variants between each SUB or SUD and each externalizing or internalizing trait. For pairs involving AFB and AFS, the direction of the genetic correlation of shared variants was reversed to indicate the genetic correlation with a higher externalizing behavior. Pairs involving SUBs (EverSmk and DrnkWk) are denoted by smaller dots, while pairs involving SUDs or SUD-proxies (Cigsperday and AUD) are represented by larger dots. Pairs involving externalizing or internalizing traits are colored in black or white, respectively. Pairs involving tobacco use behaviors or disorders (EverSmk and Cigsperday) are depicted by squares and pairs involving alcohol use behaviors or disorders (DrnkWk and AUD) are represented by triangles. ADHD, attention deficit hyperactivity disorder; AFB, age at first birth; AFS, age at first sexual intercourse; AUD, alcohol use disorder; Cigsperday, average number of cigarettes smoked per day; DEP, lifetime diagnosis of major depressive disorder or broad self-reported depression; DEP-NEURO, depressed affect subcluster of neuroticism; DrnkWk, average number of drinks drunk each week; EverSmk, ever been a regular smoker; MOOD, experiencing mood swings; NEURO, neuroticism; NSEX, lifetime number of sexual partners; RISK, general risk tolerance; SUB, substance use behaviors; SUD, substance use disorders, WORRY-NEURO, worry subcluster of neuroticism.
Among the analyzed trait pairs, a subgroup exhibited a comparable degree of genetic overlap (>90% of shared causal variants) (Figure 4). Some pairs involving SUDs and SUD-proxies with externalizing traits demonstrated a relatively high genetic correlation of shared variants (absolute values of rgs = 0.50–0.69), despite their high degree of genetic overlap, such as Cigsperday and AFB or AUD and AFS. Conversely, pairs with a similar degree of genetic overlap comprising SUDs or SUD-proxies with internalizing traits demonstrated a lower genetic correlation of shared variants (absolute values of rgs = 0.06–0.42), such as Cigsperday and WORRY-NEURO or Cigsperday and NEURO. However, it was much lower for SUBs, notably DrnkWk, and internalizing traits (absolute values of rgs = 0.02–0.16, except for EverSmk and DEP (absolute value of rgs = 0.35)).
Discussion
Using MiXeR, we detected a distinct pattern of genetic overlap between SUBs and SUDs with externalizing and internalizing traits. SUBs (i.e. EverSmk and DrnkWk) and internalizing traits demonstrated a notably high genetic overlap but a particularly low genetic correlation of shared variants. This finding may be explained by a divergence in the effect distribution of the numerous shared risk variants between SUBs and internalizing traits, suggesting a limited role in the genetics of internalizing problems at SUBs. By contrast, SUBs showed a higher genetic correlation of shared variants with externalizing traits, suggesting that externalizing problems may have a stronger genetic influence on SUBs, albeit with variations depending on the substance. With regards to SUDs or SUD-proxies (i.e. Cigsperday and AUD), we observed a notably high genetic correlation of shared variants with externalizing traits, also in cases of high genetic overlap. This observation suggests that most risk variants for SUDs and SUD-proxies are shared with externalizing problems, and these variants show a similar effect distribution on both traits. A similar pattern, although not as marked, was found for pairs involving SUDs and SUD-proxies with internalizing problems. These findings suggest that while both externalizing and internalizing pathways may be involved in SUDs, externalizing pathway seems to play a more consistent role.
Bivariate MiXeR revealed a substantial genetic overlap between SUBs and SUDs and both externalizing and internalizing traits. A similar pattern was already identified in a previous study from our group between mental disorders and other traits such as intelligence, educational attainment, neuroticism, and SWB (Hindley et al., Reference Hindley, Frei, Shadrin, Cheng, O’Connell, Icick and Andreassen2022). This study concluded that a large number of pleiotropic variants are involved in all these traits with differences in the effect size distribution of these pleiotropic variants playing a more prominent role in the genetic predisposition for a given trait than trait-specific genetic variants. Considering our results, this conclusion can also be extended to the genetic overlap of externalizing and internalizing traits with SUBs and SUDs. Our study identified two distinct patterns within this general framework. SUBs and internalizing traits showed a high genetic overlap but a genetic correlation of shared variants close to zero, suggesting a distinctly divergent effect distribution within both traits. Conversely, certain pairs involving SUDs or SUD-proxies with externalizing traits displayed a similar distribution of effects as well as a high genetic overlap, suggesting a shared genetic basis among these traits.
Our results are consistent with previous studies suggesting that, while the internalizing pathway plays a limited role at the initial stages of substance consumption, it has a stronger impact at more severe levels of use (Hussong et al., Reference Hussong, Jones, Stein, Baucom and Boeding2011; Ning et al., Reference Ning, Gondek, Patalay and Ploubidis2020). A different genetic relationship of internalizing traits with SUBs and SUDs has already been suggested using MiXeR (Icick et al., Reference Icick, Shadrin, Holen, Karadag, Lin, Hindley and Andreassen2022). In this previous study, mood instability showed a different pattern of genetic overlap with alcohol consumption, defined as AUDIT-C, and AUD. Here, we have expanded on these findings by using several internalizing traits in addition to mood instability and a different alcohol consumption trait (DrnkWk). Our findings are also consistent with previous studies suggesting a role of the externalizing pathway at all substance use stages. An effect of the externalizing pathway on substance use initiation was suggested in a previous study which found that the shared genetic susceptibility between schizophrenia and smoking initiation is related to externalizing behavior (Al-Soufi & Costas, Reference Al-Soufi and Costas2023). Importantly, our results emphasize the substantial role of the externalizing pathway in SUDs, consistent with previous studies supporting a clearer association of externalizing problems with severe substance use compared to substance consumption and internalizing problems (Edwards, Gardner, Hickman, & Kendler, Reference Edwards, Gardner, Hickman and Kendler2016; Ning et al., Reference Ning, Gondek, Patalay and Ploubidis2020). Previous studies have implicated the genetic susceptibility to both externalizing and internalizing problems in SUDs. A recent study found that an addiction risk factor based on problematic alcohol use, problematic tobacco use, CUD, and opioid use disorder GWAS correlated genetically with externalizing behavior (absolute rg = 0.35–0.52), as well as with substance use to cope with anxiety (rg = 0.63) and depression (rg = 0.60), suggesting a strong association of externalizing and internalizing behavior with SUDs (Hatoum et al., Reference Hatoum, Colbert, Johnson, Huggett, Deak, Pathak and Agrawal2023). Another study based on a similar addiction risk factor found that the association between this addiction risk factor and risk-taking significantly decreased after taking substance use into account (Hatoum et al., Reference Hatoum, Johnson, Colbert, Polimanti, Zhou, Walters and Agrawal2021). In contrast, genetic susceptibility to substance use had a small effect on the association between this addiction risk factor and neuroticism. Our findings are compatible with these results suggesting that, in contrast to the internalizing pathway, the effect of the externalizing pathway on SUDs may be partially mediated by substance use.
For SUDs and SUD-proxies, both tobacco and alcohol showed comparable patterns of genetic overlap with externalizing and internalizing traits. However, differences emerged between the tobacco (EverSmk) and alcohol (DrnkWk) use behaviors. EverSmk displayed a higher genetic correlation of shared variants with externalizing and internalizing traits compared to DrnkWk. Moreover, the genetic correlation of shared variants of internalizing traits with DrnkWk was close to 0, suggesting that the internalizing pathway may have a minimal effect on alcohol use. In contrast to tobacco use, DrnkWk is a highly heterogenous trait, combining the frequency of use and quantity per drinking occasion (Mallard et al., Reference Mallard, Savage, Johnson, Huang, Edwards, Hottenga and Sanchez-Roige2022; Sanchez-Roige et al., Reference Sanchez-Roige, Cox, Johnson, Hancock and Davis2021). These dimensions show opposite genetic correlations with traits related to socioeconomic status, psychiatric disorders, and psychological traits (Marees et al., Reference Marees, Smit, Ong, Macgregor, An, Denys and Derks2019). As a consequence of this heterogeneity, DrnkWk is expected to be less influenced by the internalizing and externalizing pathways than use of tobacco or other substances.
The present study has some limitations. First, we cannot conclude that there is a causal effect of externalizing and internalizing problems on SUBs and SUDs on the basis of MiXeR results alone. We can only assess whether there is a shared genetic susceptibility between both traits and its effect distribution on both traits. Second, some of the MiXeR models were not distinguishable from a complete genetic overlap or the minimum genetic overlap allowed for a given genetic correlation. Therefore, larger GWAS based on a more precise phenotype definition is required to achieve more accurate estimates of the number of shared and unique genetic variants. Third, a lack of well-powered GWAS for other substances only allowed the analysis of substance use for two legal substances: alcohol and tobacco. Thus, our conclusions may not generalize to other substances. Fourth, phenotype definition remains a major topic in psychiatric genetics. A continuous trait for maladaptive alcohol use would assess more accurately the level of substance dependence of an individual. Adding data regarding the course of SUD (age at onset, continuous versus discontinuous use, duration of SUD, speed, and age of transition between use and SUD) may be relevant in refining the genetic component of SUD. This limitation of the dichotomous approach is extensive to other traits used in this study. Furthermore, differences in the procedures used to ascertain cases may be associated with measurement bias. In general, there is a need for more comprehensive assessments of GWAS samples regarding traits of relevance in psychiatry. Unfortunately, most of the phenotypic refinements susceptible to improved transfer to biology or the clinics occur at the expense of sample size, thus reducing statistical power. Fifth, there is considerable comorbidity between externalizing and internalizing behavior, which makes it difficult to distinguish the separate effects of each analyzed pathway. Moreover, many GWAS rely on samples like UK Biobank, which may not accurately represent the general population in terms of gender, socio-economic status, or ancestry, potentially biasing our results (Fry et al., Reference Fry, Littlejohns, Sudlow, Doherty, Adamska, Sprosen and Allen2017). In addition, since MiXeR assumes that the effect size of causal variants is normally distributed, the cluster of nAchR genes at chromosome 15 was excluded from our Cigsperday GWAS. MiXeR also assumes an additive model of genetic effects, while gene–environment interactions, epistasis, and dominance effects may play an important role, particularly in personality traits such as neuroticism (Boomsma et al., Reference Boomsma, Helmer, Nieuwboer, Hottenga, de Moor, van den Berg and de Geus2018; Keller, Coventry, Heath, & Martin, Reference Keller, Coventry, Heath and Martin2005). Violation of the MiXeR’s assumption of additivity may therefore contribute to the weak genetic correlations with neuroticism. Furthermore, we did not provide statistical significance for our results, as formal significance tests based on MiXeR results are not feasible. Finally, although we used several sexual behaviors as representative traits of genetic susceptibility for externalizing behavior, these complex phenotypes are affected by gene–environment correlations attributable to social, economic, and political processes (Abdellaoui, Dolan, Verweij, & Nivard, Reference Abdellaoui, Dolan, Verweij and Nivard2022). Therefore, a cautionary remainder is needed to avoid misinterpretation of genomic findings that may lead to stigmatization.
In summary, we found a different genetic relationship between SUBs and SUDs with externalizing and internalizing traits, suggesting a larger involvement of the externalizing and internalizing pathways in severe substance use. In addition, the externalizing pathway seems to play a more prominent role throughout the progression of substance use than the internalizing pathway. These findings improve our understanding of the origins of substance use and SUDs, which may inform attempts to advance prevention and treatment strategies.
Supplementary material
The supplementary material for this article can be found at http://doi.org/10.1017/S0033291725000108.
Acknowledgements
The authors wish to thank all participants in the studies included in this paper and all Consortia and research groups providing the summary statistics datasets used in this work. Specifically, we would like to thank the GWAS and Sequencing Consortium of Alcohol and Nicotine use, the Psychiatric Genomics Consortium, the Social Science Genetic Association Consortium, the Complex Traits Genetics Lab, the UK Biobank, the International Cannabis Consortium, the Early Genetics and Lifecourse Epidemiology, the Million Veteran Program, the Neale lab, and the GWAS Catalog. This work was performed on the Galicia Supercomputing Center (CESGA) facilities and the TSD (Tjeneste for Sensitive Data) facilities, owned by the University of Oslo, operated and developed by the TSD service group at the University of Oslo, IT-Department (USIT) ([email protected]). This research has been conducted using data from UK Biobank, a major biomedical database (www.ukbiobank.ac.uk).
Funding statement
This work was supported by the Instituto de Salud Carlos III (grant number RIAPAd RD21/0009/0011). LA-S was supported by a research fellowship from the Ministerio de Universidades (grant number FPU18/03243). We gratefully acknowledge support from the Research Council of Norway (RCN) (296030, 324252, 324499, 300309, 273291, 223273), the South-East Norway Regional Health Authority (2022–073), and KG Jebsen Stiftelsen (SKGJ-MED-021). This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreements No 847776 and 964874.
Competing interests
OAA is a consultant to cortechs.ai and Precision Health, and received speaker’s honorarium from Lundbeck, Otsuka, Janssen and Sunovion unrelated to the topic of the current study.








