- DC
dendritic cells
- MG
mammary gland
- MLN
mesenteric lymph nodes
- PBMC
peripheral blood mononuclear cells
- rDNA
ribosomal DNA
- TP
time point
- TTGE
temporal temperature gradient-gel electrophoresis
Understanding of the association and interactions between the intestinal microbiota and the host immune system has advanced greatly in recent years. However, mammals are born sterile and the events shaping the establishment of a flourishing intestinal microbiota remain elusive. It is known that dendritic cells (DC), via pattern recognition receptors such as toll-like receptors, sample the microbes that continuously bombard the intestinal mucosa(Reference Stagg, Hart and Knight1). The end result is tolerance to the normal microbiota and protection against pathogenic attack. Yet, successful, simultaneous deployment of such divergent processes requires highly sophisticated control mechanisms which are not expected of an inexperienced neonatal immune system. The very survival of the neonate after the massive bacterial insult in the birth canal is testimony to solid regulatory mechanisms which may already operate during fetal life. The subsequent assembly of specific bacterial communities, in the absence of adverse immune responses, is testimony to its long-term effects and robustness. Environmental factors, including the method of delivery and the use of antibiotics, impact the final composition of the neonatal microbiota(Reference Falk, Hooper and Midtvedt2). However, differences between breast-fed and formula-fed infants in intestinal bacterial colonization(Reference Falk, Hooper and Midtvedt2) and in susceptibility to intestinal disease or sepsis(Reference Wright, Bauer and Naylor3) suggest that further education is also acquired by factors in breast milk.
More specifically, breast milk provides the suckling infant with a myriad of soluble factors such as Ig, lactoferrin and lysozyme, which influence the composition of the neonatal microbiota and play a direct role in the protection of the neonatal intestine(Reference Lonnerdal4). Another possibility is that soluble pattern recognition receptors such as CD14 and toll-like receptor 2 present in milk have an indirect effect by instructing the neonatal immune system to recognize and respond appropriately to commensals and pathogens(Reference Labeta, Vidal and Nores5, Reference Vidal, Donnet-Hughes and Granato6). However, there is accumulating evidence that bacteria of maternal origin are transmitted to the infant via colostrum and milk(Reference Moughan, Birtles and Cranwell7). The majority of milk organisms arise from the mother's skin or the infant's mouth(Reference Moughan, Birtles and Cranwell7, Reference Gavin and Ostovar8), but certain species are suggested to colonize the neonatal intestine(Reference Martín, Langa and Reviriego9). Most studies on the microbiology of human milk have addressed the transmission of pathogens or contaminating commensals in samples meant for milk banks(Reference Gavin and Ostovar8, Reference West, Hewitt and Murphy10, Reference El Mohandes, Schatz and Keiser11). However, it has been suggested that breast milk is not sterile, even when collected aseptically(Reference West, Hewitt and Murphy10). This raised the interesting possibility that breast milk harbours a natural bacterial inoculum.
Here, we examined whether some microbial species are indeed intrinsic to human milk. Confirmation of a bacterial inoculum, albeit of low dose, prompted a subsequent investigation into its origin. Since human breast milk leucocytes arise from cells which have migrated from the intestine to the lactating mammary gland (MG) via the lymphatics and the circulation, we considered that this cellular circuitry conveys microbial components in a manner which is not deleterious to maternal health. Consequently, we examined the bacterial profile associated with the cellular compartments of human milk and maternal blood and compared this to the profile in maternal and infant faeces. This was followed by a study of bacterial colonization in various tissue compartments of virgin, pregnant and lactating mice. Taken together, our results suggest that bacterial translocation is a physiological process which is augmented in the pre- and postpartum period, one of the consequences of which is the cellular transfer of bacterial components to the suckling infant via the breast milk. The biological relevance of these phenomena to the mother and to the suckling infant remain a matter of conjecture. However, if one considers the risks associated with such an evolutionary process, it is likely that the motive goes beyond simple colonization of the neonatal intestine by small numbers of milk-borne organisms.
Methods
Human milk, blood and faecal samples
Breast milk was collected from healthy lactating mothers who delivered at term. Foremilk was discarded, the breast cleaned with antiseptic soap, rinsed with sterile distilled water and dried using sterile gauze before aseptic collection using an electric breast pump. As a control, a swab of the areola was taken before milk collection. Samples of whole milk were plated on Man, Rogosa and Sharpe medium containing cysteine, on Eugon Tomato, Drigalski, Shaedler Neo Vanco or on blood agar (Bio-Mérieux, Marcy l'Etoile, France) and incubated aerobically or anaerobically at 37°C. Cells were collected from the remaining milk by centrifugation and exposed to sterile PBS containing 1% gentamycin to kill extracellular bacteria. Cells were aliquoted and used to make cytopreparations, frozen in RPMI media (Life Technologies, Basel, Switzerland) containing 10% dimethylsulphoxide in fetal calf serum for flow cytometric analysis or lysed using cold sterile distilled water and passed through a sterile needle for plating on bacterial culture media.
Venous blood (10 ml) was collected from eight lactating women at different times postpartum or from five age-matched, non-gravidus, non-lactating women. The blood was centrifuged over Ficoll-Hypaque media (Sigma-Aldrich, St. Louis, MO, USA), washed and then processed as for milk cells. Maternal and infant faecal samples were collected in sterile tubes, aliquoted and stored frozen at −80°C until required. Protocols were approved by an internal ethical committee and the Swiss authorities.
Flow cytometry
Myeloid and lymphoid DC in the peripheral blood mononuclear cells (PBMC) fraction were examined using the FACSCalibur and DC-KIT (Becton Dickinson, Basel, Switzerland). This kit contains fluorescein isothiocyanate-conjugated lineage cocktail with antibodies to CD3, CD14, CD16, CD19, CD20 and CD56; peridinin chlorophyll protein-conjugated anti-human leucocyte antigen-DR, allophycocyanin-conjugated anti-CD11c and phycoerythrin-conjugated anti-CD123. In separate tubes, cells were labelled with fluorescein isothiocyanate-anti-CD11c and PE-anti CD14 (Becton Dickinson) according to the manufacturer's instructions before analysis using the FACSCalibur.
Temporal temperature gradient gel electrophoresis
Total DNA was extracted from 200 mg faecal samples and from milk and blood samples as described previously for faeces and biopsies, respectively(Reference Lepage, Seksik and Sutren12), except that for milk and blood, DNA precipitations were done overnight and the pellets centrifuged for 1 h at 4°C.
Isolated DNA were then used as templates to amplify the V6–V8 regions of 16S ribosomal DNA (rDNA), using primers U968-GC-F and L1392-R(Reference Lepage, Seksik and Sutren12). Several dilutions of template DNA were tested if the presence of PCR inhibitors was suspected, in order to optimize PCR yield and limit possible bias. The PCR amplification and temporal temperature gradient-gel electrophoresis (TTGE) were performed as before(Reference Mangin, Bonnet and Seksik13) and Gel Compar II software (Applied Maths) used to compare TTGE profiles. A marker consisting of a PCR amplicon mix of seven cloned rDNA from different bacterial species was used to normalize the profiles.
DNA from milk, maternal blood cells and infant faecal samples at week 4 postpartum were amplified by PCR with primers U350-F and L1392-R. Ligation and cloning in pGEM-T Vector system I (Promega) were performed as described previously(Reference Mangin, Bonnet and Seksik13). Forty-eight clones per library were sequenced using the primers M13F and M13R. An equal portion of the small subunit rDNA (Escherichia coli position 350 to 1392) was used for the sequence analysis. These sequences were checked manually and contig were made using BioEdit software. These sequences were submitted to Genbank for preliminary analysis using the Blast programmes (Blast and Megablast) of the Ribosomal Database Project in order to identify putative, close phylogenetic relatives. The sequences were tested for chimera structure using the Ribosomal Database Project analysis service Check Chimera and during manual inspection of alignment. The sequences were aligned to their nearest neighbour with the automated alignment tool of the ARB programme package and the arb database (Technical University of Munich) of January 2004. Mother and infant sequences were compared using the Blast2sequences programme of the National Center for Biotechnology Information.
Bacteria localization in human milk and blood cells
After fixation in absolute ethanol, cytopreparations of human milk and blood cells were incubated for 5 min with 100 mg/ml acridine orange(Reference Ho and Lawton14, Reference Kite, Millar and Gorham15), washed extensively, mounted in fluorescent mounting medium (DakoCytomation, DAKO Schwiez, Baar, Switzerland) and analysed by epifluorescence microscopy.
IL-12 and fetal liver tyrosine kinase-3 ligand
IL-12 and fetal liver tyrosine kinase-3 ligand in human serum (1:2 dilution) were assayed by ELISA according to the manufacturer's instructions (Diaclone and R&D Systems, Epalinges, Switzerland).
Animal study
Conventional virgin and pregnant/lactating C57BL/6 mice (Charles River, l'Arbresle, France) were sacrificed (n 10 per group) at 5–6 d before parturition, or at 1–2, 3–4 or 14–15 d postpartum. Samples of blood, intestinal contents, mesenteric lymph nodes (MLN), spleen, liver and MG were aseptically collected for microbiological analysis, fixed in Bouin's fixative before mounting in paraffin blocks and/or mounted in optimal cutting temperature compound and frozen in liquid nitrogen. The experimental procedure was approved by an internal ethical committee and the Swiss authorities.
Bacteria in mouse tissues
Micro-organisms were visualized in tissues using Gram's stain or a rabbit antibody to lipoteichoic acid(Reference Granato, Bergonzelli and Pridmore16) and tetramethylrhodamine β-isothiocyanate-labelled anti rabbit-IgG (Jackson) as secondary antibody. For microbiological analysis, samples of mouse tissues were homogenized, suspended in sterile PBS, plated onto blood agar (Bio Mérieux) and incubated aerobically or anaerobically at 37°C.
Fluorescent labelling of mouse tissues
Thin sections (5 μm) of optimal cutting temperature compound-embedded tissues were fixed in cold ethanol and rehydrated using PBS. Non-specific staining was blocked using 10% fetal calf serum in PBS before labelling with fluorescein isothiocyanate-labelled hamster anti-mouse CD11c or the control isotype hamster IgG1 (Becton Dickinson) according to the manufacturer's instructions.
Statistical analysis
The proportion of animals with bacteria in different tissues was analysed by the Fisher's exact test. Differences in the median percentage of DC in human blood were compared using the Mann–Whitney test. TTGE profiles were compared using Gel Compar II software (Applied Maths, Kortrigk and Belgium). Similarity coefficients (Pearson correlation method) were then calculated for each pair of profiles, yielding a similarity matrix. A dendrogram was constructed from this matrix by using an Unweighted Pair Group Method with Arithmetic Mean algorithm.
Results
Bacterial signatures common to the neonatal microbiota and maternal blood and milk cells
To minimize contamination of expressed milk with skin bacteria or organisms residing in the superficial ducts of the breast, the foremilk was rejected and the breast cleaned with antiseptic soap prior to milk collection. No bacteria were grown from swabs of the nipple areola. Expressed milk contained a total concentration of <103 cfu/ml bacteria composed of Lactobacillus, Streptococcus, Enterococcus, Peptostreptococcus, Staphylococcus, Corynebacterium and/or an occasional Escherichia species.
Since endogenous intracellular trafficking of bacterial components from the intestine to the lactating breast would implicate the lymphatic and systemic circulation, we next used TTGE to examine the bacterial rDNA content in human milk cells and maternal PBMC and faeces during lactation. Furthermore, we examined corresponding infant faecal samples to address a possible transmission of maternal bacteria via milk. The advantages of TTGE analysis lie in its sensitivity and its capacity to detect non-culturable bacteria. Thus, a more extensive number of bacteria can be catalogued.
Maternal faecal samples gave classic TTGE profiles which were specific for each individual and of greater biodiversity than those of infant faeces (Fig. 1). Although breast milk cells harboured a less complex microbiota than in maternal faeces, TTGE revealed a greater biodiversity than previously observed by plating of the milk. Thus, non-culturable bacteria or the DNA from dead bacteria may also be present intracellularly. Fig. 1 shows one mother–infant couple analysed over weeks 1–4 postpartum. Similar TTGE profiles were observed for seven other mother–infant pairs (data not shown). Interestingly, some bacterial signatures (indicated by arrows) were common to the infant faeces and to several samples of maternal origin (Fig. 1). Upon excision from the TTGE gel and sequencing, the lowest of these bands, especially intense in infant faeces and comigrating in maternal faeces and blood, was identified as Bifidobacterium longum on the basis of 369 nucleotides. Sequencing of the other common bands identified DNA from Streptococcus salivarius and Rhodococcus erythropolis.

Fig. 1. Transmission of bacterial signatures from mother to child via maternal peripheral blood mononuclear cells and milk cells during lactation. Profiling of dominant bacterial signatures in maternal faeces, blood and milk cells and in infant faeces using temporal temperature gradient-gel electrophoresis amplified fragments of ribosomal DNA (rDNA). Each dominant bacterial species leads to the formation of one or a few bands in the pattern. Duplicate PCR were used for the maternal blood and milk cell fractions. A ladder (L) obtained by PCR amplification of cloned rDNA sequences was used for normalization of gels and image analysis. Some bacterial signatures (indicated by arrows) were common to the infant faeces and to samples of maternal origin.
Next, PCR products were used to prepare rDNA libraries from the milk cells. Besides the species previously identified by plating and by sequencing of the excised TTGE gel bands, sequencing of milk cell clones revealed the presence of Bacteroides, Clostridia, Eubacterium and Enterobacteria among a total of 15 genera typical of ileal and colonic microbiota. The most represented bacterial genera in infant faeces were less diverse and essentially lactose degrading, lactic acid producing bacteria together with Staphylococcus species. Out of 23 sequences corresponding to bifidobacteria, 17 were related to B. longum, five to Bifidobacterium bifidum and 1 to Bifidobacterium infantis. Two identical rDNA sequences (99% identity out of 1117 base pairs), corresponding to Streptococcus thermophilus and Staphylococcus epidermidis, were identified in the milk cell clones and in the infant's faeces (data not shown).
Interestingly, PBMC of lactating mothers contained a restricted variety of bacterial rDNA sequences (Fig. 1). Bacterial signals were also present in blood cells of non-gravidus, non-lactating women, but the complexity of bacterial signatures was greater in PBMC from breast-feeding mothers (data not shown). Profiles for control women were similar, while those of lactating women were specific for each individual.
Acridine orange staining of milk and blood cytopreparations identified bacterial bodies in association with mononuclear cells (Fig. 2).

Fig. 2. Intact bacterial structures in milk cells and peripheral blood mononuclear cells (PBMC) from lactating mothers stained with acridine orange.
Increased translocation of intestinal bacteria occurs in pregnant and lactating mice
To further define endogenous cellular transport of bacterial components, we next examined intestinal uptake of bacteria in conventional non-pregnant, pregnant and lactating mice.
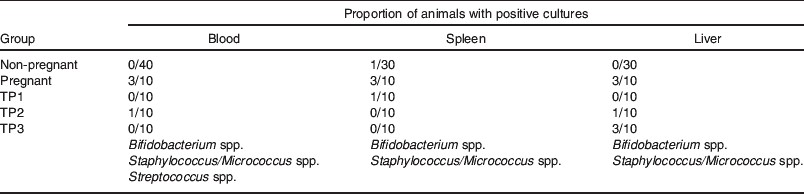
In contrast to the confined bacterial translocation to MLN observed in control animals, heightened translocation to MLN in the perinatal period was followed by bacterial colonization of the breast in the immediate postpartum period (Fig. 3). While 10% control animals (C) had positive MLN cultures, 70% pregnant animals had detectable levels of bacteria in their MLN, 5–6 d before delivery (time point (TP) 1). Within 24 h postpartum, fewer animals had positive MLN cultures, but 80% of the mice had viable bacteria in the mammary tissue. Although this value decreased to 50% by 3–4 d postpartum, it was still significantly different from that of control mice (P<0·005). From the present study, one cannot deduce whether subsequent waves of bacteria translocate at other time points later during lactation. Bacterial colonization of the MG coincided with an increased number of positive blood cultures and occasional translocation to the spleen and liver (Table 1). In contrast to pathological conditions where translocating micro-organisms are mainly Gram negative, penetrating organisms in pregnant and lactating mice included Streptococcus, Lactobacillus and Bifidobacterium species whose numbers gradually subsided over time (data not shown).

Fig. 3. Microbiological analysis of mouse tissues reveals increased bacterial translocation during pregnancy and lactation. Proportion of control virgin mice (C), pregnant mice (5–6 d before parturition, TP1) and lactating mice at 1–2 d (TP2), 3–4 d (TP3) and 14–15 d (TP4) postpartum with viable bacteria in mesenteric lymph nodes (MLN) (![]() ) and mammary gland tissue (MG) (□). ***P<0·00005; **P<0·005; *P<0·05 as compared to controls (Fisher's exact test).
) and mammary gland tissue (MG) (□). ***P<0·00005; **P<0·005; *P<0·05 as compared to controls (Fisher's exact test).
Table 1. Bacterial translocation to extra-intestinal tissues during pregnancy and lactation. The proportion of animals with viable bacteria in the spleen, liver and blood was examined after plating on blood agar. Positive samples had a low number of micro-organisms (blood: 101–102 cfu/ml; spleen and liver: 102–103 cfu/g). Bacteria isolates were characterized by macroscopic and microscopic morphology, Gram's staining, culture characteristics and using Fluorescence in situ hydridisation technology(Reference Perez, Doré and Leclerc42) and specific probes (Bif 1412, MWG Biotech). Time points TP1, TP2 and TP3 are the same as for Fig. 3.
During lactation, bacteria were visualized histologically in the lamina propria of the small bowel, in the sub-epithelial dome and interfollicular regions of the Peyer's patch and associated with cells in the glandular tissue of the MG (data not shown). Double labelling using antibodies to CD11c and bacterial lipoteichoic acid showed co-localization of micro-organisms and CD11c+ cells suggesting that bacterial cells were inside the DC (data not shown).
The Peyer's patches of pregnant and lactating mice were enlarged compared to those of controls, had a more prominent sub-epithelial dome and more dilated lymphatic vessels (Fig. 4). Furthermore, CD11c+ cells were localized predominantly in the sub-epithelial dome and interfollicular regions of the Peyer's patches. In the patches of pregnant and lactating animals, mononuclear cells were observed within the draining lymphatics (Fig. 4).

Fig. 4. Staining of Peyer's patch tissue from a pregnant mouse. Haemotoxylin and eosin staining showed a reactive lymphoid tissue with dilated lymphatic vessels containing mononuclear cells (lower left, ×100 and lower right, ×1000) that are not present in virgin mice (upper left, ×100). Fluorescently labelled CD11c+ cells were prominent in the sub-epithelial dome and interfollicular region (upper right).
Myeloid and lymphoid dendritic cells are diminished in the circulation of lactating women
Breast milk has a high proportion of phagocytes which readily ingest microbes but are ineffective at killing(Reference Buescher and McIlheran17). Recent work demonstrating that CD14+ milk mononuclear cells express HLA-DR, CD86 and CD83 as well as DC-specific intercellular adhesion molecule 3-grabbing non-integrin, a C-type lectin considered to be exclusively expressed on DC, suggests that these cells are partially differentiated DC(Reference Ichikawa, Sugita and Takahashi18). Since milk DC-like cells may be derived from the maternal circulation and are potential vehicles of intestinally derived microbial components, we next evaluated the distribution of DC phenotypes in the PBMC of lactating and non-lactating women using flow cytometry. The percentages of CD123+ lymphoid DC and CD11c+ myeloid DC were determined after gating on lin− HLA-DR+ PBMC (data not shown). In addition, the numbers of CD14+CD11c+, potential DC precursors were studied. We found that the frequencies of differentiated lymphoid DC (lin−CD14−HLA-DR+CD11c−CD123+) and myeloid DC, (lin−CD14−HLA-DR+CD11c+CD123−) phenotypes and CD14+CD11c+ cells in the circulation of lactating women are lower than those of controls. Of note, both CD14hiCD11c+ and CD14loCD11c+ cells are present in the circulation of control and lactating women and while there is a tendency towards a higher proportion of the CD14loCD11c+ intermediate DC-like phenotype during the first week of lactation, the results were not significantly different from controls.
Discussion
After discarding the foremilk, we found that aseptically collected human breast milk contained a low total concentration of up to 103 cfu/ml microbes comprising Lactobacillus, Streptococcus, Enterococcus, Peptostreptococcus, Staphylococcus and/or Corynebacterium, with an occasional Escherichia species. In spite of every precaution, some of these isolates may still arise from contamination. One might argue that bacterial contaminants in milk may be biologically relevant to the breast-fed infant. Indeed, Staphylococcus species are common constituents of the early neonatal microbiota(Reference Falk, Hooper and Midtvedt2). However, a bacterial presence in all samples suggests that a discrete microbiota may naturally exist in breast milk. Consistent with this, we made the observation that the bacteria were associated with breast milk cells examined microscopically (Fig. 2).
Human breast milk contains an appreciable leucocyte population, the lymphocytes of which originate from the gut-associated lymphoid tissue and bronchial lymphoid tissue of the mother(Reference Roux, McWilliams and Phillips-Quagliata19) and the mononuclear phagocytes from peripheral blood(Reference Goldman and Goldblum20). Here, we propose that the latter also arise in the gut-associated lymphoid tissue, capture components of the luminal microbiota prior to their departure and transfer them to the suckling infant via breast milk. Since it is generally accepted that tissue macrophages are resident, non-migrating cells, we considered DC-like cells as the most likely vehicles for trafficking within the common mucosal system.
In contrast to the two or three viable bacterial genera observed by plating of individual milk samples, TTGE analysis of human milk cells revealed a wide diversity of bacterial signatures, corresponding to the dominant autochthonous ileal and colonic commensal micro-organisms. This confirms the expected uptake of bacteria at these tissue sites and suggests that non-culturable bacteria or the DNA from dead bacteria may also be present intracellularly.
Migration of bacteria, or bacterial components, within intestinally derived leucocytes to the lactating breast is supported by the observation that some of the rDNA bands on TTGE gels are common to the faecal, blood and milk samples of each mother examined. Furthermore, some of these bands are also found in infant faeces (Fig. 1) and may represent microbes transferred to the suckling infant via the milk.
To our surprise, PBMC contained a restricted variety of bacterial rDNA sequences which was more extensive in the blood of breast-feeding mothers. However, no viable bacteria were isolated from these cells. The reason for this discrepancy is not known. One could speculate that the few, bacterially laden cells may be diluted in the circulation. However, it is also possible that certain blood-borne bacteria may be quiescent or dead due to intracellular antimicrobial effects.
Awareness that some bacteria may nevertheless be environmental contaminants and that PCR amplification may have inadvertently led to false positives, we confirmed microscopically whether bacteria were associated with cells in the milk or maternal blood. Unlike sepsis where translocating bacteria are associated with polymorhonuclear cells(Reference Kite, Millar and Gorham15), intact bacterial bodies were observed in association with a limited number (<0·1%) of milk and blood mononuclear cells (Fig. 2). However, it is not excluded that the components from dead organisms may be associated with milk and blood polymorphonuclear cells.
During lactation, bacteria were visualized beneath the epithelium of both the Peyer's patch dome and the intestinal lamina propria of mice. This suggests M-cell-mediated uptake towards DC in the Peyer's patches, direct sampling of luminal bacteria by dendrites of lamina propria DC or a low-level, physiological leakiness of the epithelium(Reference Uhlig and Powrie21). In any event, an extremely limited number of bacteria are thought to cross the intestinal epithelium, evade uptake and killing by intestinal macrophages, but remain viable after phagocytosis by DC(Reference MacPherson and Uhr22). Bacterially loaded DC are then thought to migrate to the MLN where they initiate protective immune responses(Reference MacPherson and Uhr22). These migrating DC phagocytose bacteria well but are relatively ineffective at killing internalized organisms(Reference Nagl, Kacani and Mullauer23). The more prominent Peyer's patches observed in pregnant and lactating animals, with localization of CD11c+ at the sites of noticeable mononuclear cell exit through lymphatic vessels (Fig. 4), provides indirect support of mononuclear cell-mediated transport of intestinally derived microbes to the lactating MG.
The presence of a modest population of enteric-like bacteria in association with a milk cell compartment suggests that this inoculum might serve an alternative purpose to that traditionally associated with bacterial implantation in the neonatal intestine. These organisms may indeed eventually colonize neonatal tissues, but we speculate that they educate the neonatal immune system in the recognition of commensal-associated molecular patterns of bacteria.
It has been previously shown that human milk contains soluble pattern recognition receptors for bacterial molecular motifs and that these may mediate a differential response to Gram-negative and Gram-positive organisms(Reference Labeta, Vidal and Nores5, Reference Vidal, Donnet-Hughes and Granato6). Uptake of maternal leucocytes into neonatal tissues occurs during gestation and lactation(Reference Zhou, Yoshimura and Huang24). Perhaps prolonged penetration of inconspicuous bacterial molecular patterns via maternal DC, induces tolerogenic responses that are analogous to those for self-antigens. The TNF-family molecule receptor activator of NF-κB ligand (osteoprotegerin ligand, TNF-related activation-induced cytokin and osteoclast differentiation factor) and its receptor activator of NF-κB are key regulators of bone remodelling(Reference Theill, Boyle and Penninger25) and regulate T-cell/DC differentiation communication(Reference Anderson, Maraskovsky and Billingsley26, Reference Josien, Wong and Li27), lymph node formation(Reference Kong, Yoshida and Sarosi28) and the development of a lactating MG during pregnancy(Reference Fata, Kong and Li29). It is noteworthy that osteoprotegerin, a soluble decoy receptor for receptor activator of NF-κB ligand, is a DC survival factor(Reference Wong, Josien and Lee30) which is elevated in serum during pregnancy and lactation(Reference Uemura, Yasui and Kiyokawa31) and is present in significant quantities in human breast milk(Reference Vidal, van den Broek and Lorget32). It has been shown that Peyer's patch DC express receptor activator of NF-κB and that treatment with receptor activator of NF-κB-Fc, which like osteoprotegerin, antagonizes receptor activator of NF-κB ligand activity and enhances the induction of tolerogenic immune responses(Reference Williamson, Bilsborough and Viney33). It is feasible that the high levels of osteoprotegerin during pregnancy and lactation influence the generation of tolerogenic v. activating DC in the mother. It is also possible that milk osteoprotegerin has a similar effect in the suckling neonate.
Elevated translocation of bacteria or their components in the mother should certainly have some bearing on her immune status. The traditional paradigm that normal pregnancy is a T-helper 2-mediated phenomenon is challenged by reports of physiological activation of innate immunity with monocyte and granulocyte activation and increased production of IL-12 and TNF-α(Reference Sacks, Redman and Sargent34–Reference Kupferminc, Peaceman and Wigton36). We consistently observed IL-12 in the serum of three out of eight lactating mothers (range 12·8–27·3 pg/ml, data not shown). An increased physiological translocation of viable and dead intracellular bacteria or their constituents could explain this activation. Interestingly, cytosine-phosphorothioate-guanine oligodeoxynucleotides stimulate innate immunity in pregnant mice, improve maternal survival and prevent pathogen transmission to the fetus(Reference Ito, Ishii and Shirota37). The association between specific intra-uterine infections or exaggerated production of T-helper 1 cytokines, and the risk for spontaneous premature birth or abortion may reflect dysregulation of the quantity or type of microbial components translocating.
We detected a decrease of DC subsets in the blood of lactating women. These findings are in agreement with previously reported results that showed reduced numbers of circulating DC subsets in the third trimester of pregnancy(Reference Ueda, Hagihara and Okamoto38). These decreased DC numbers may reflect cellular trafficking towards the breast or intestine. Indeed, we detected fetal liver tyrosine kinase-3 ligand, a stimulator of DC differentiation and mobilization(Reference Maraskovsky, Brasel and Teepe39), in serum samples of three out of eight mothers (range 13·9–71·7 pg/ml, data not shown).
Our observations not only imply a novel form of mother–infant communication, but they also highlight a potentially new mechanism of immune regulation in healthy individuals. As shown previously(Reference Nikkari, McLaughlin and Bi40, Reference McLaughlin, Vali and Lau41), the blood of normal healthy subjects contains bacterial components. Some DNA may arise from human or microbial contamination. However, the greater number of bacterial DNA signatures in the PBMC of healthy, lactating women suggests that certain bacterial species are inherent to circulating cells. It is tempting to speculate that this represents an evolutionary strategy of immune surveillance and that such bacterial imprinting maintains tolerance to specific bacterial species and alerts distant anatomical sites of changes in local lymphoid tissues. Such a mechanism may also provide a colonization advantage to bacteria of the mother's intestinal microbiota at a time when the low bacterial diversity in the neonatal intestine is permissive to colonization.
Acknowledgements
A. D.-H., J. B., P. S., I. S.-R. and E. J. S. are Nestlé employees. P. F. P., J. D., M. L. and F. L. declare no conflict of interest. P. F. P. performed animal studies, microbiology, analysis of bacteria-associated human cells and flow cytometry; J. D., M. L. and F. L. performed TTGE analysis and cloning of bacterial DNA; J. B. contributed to the animal studies and cytokine analysis in human blood; P. S. and I. S. R. prepared and analysed samples for flow cytometry and immunohistochemistry; E. J. S. and A. D. H. generated the original hypothesis, prepared and analysed human samples, contributed to the animal studies, histological and cell analysis. A. D. H. and E. J. S. wrote the manuscript. All the authors discussed the results and commented on the manuscript. Much of the work described in this paper has been published elsewhere(Reference Perez, Doré and Leclerc42).







