Introduction
Pearl millet [Pennisetum glaucum (L.) R. Br.] is a food security crop in the harsh drylands of Sahelian West and Central Africa (WCA) and in drylands of South Asia, especially India. Additionally, farmers use the stover as animal fodder and building material.
West Africa (WA) is a primary diversity centre of pearl millet (Brunken et al., Reference Brunken, de Wet and Harlan1977; Oumar et al., Reference Oumar, Mariac, Pham and Vigouroux2008; Tostain and Marchais, Reference Tostain, Marchais and Serge1993). The International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) is maintaining a collection of 22,888 accessions of the genus Pennisetum from 51 countries (Upadhyaya et al., Reference Upadhyaya, Reddy and Gowda2007) with 13,185 accessions originating from Africa (Genesys, 2016). Characterization and agronomic evaluation of genetic resources is a prerequisite to identify trait-specific germplasm for crop improvement. Continuous, targeted diversification of breeding populations is important to assure long-term selection gains.
Pucher et al. (Reference Pucher, Sy, Angarawai, Gondah, Zangre, Ouedraogo, Sanogo, Boureima, Hash and Haussmann2015) studied 360 pearl millet accessions from 16 WCA countries at six locations in WA. Reported ranges for 13 agro-morphological traits reflected the tremendous pearl millet diversity in WCA. Bashir et al. (Reference Bashir, Ali, Ali, Melchinger, Parzies and Haussmann2013) found large variability for iron (20–86 mg/kg) and zinc (14–82 mg/kg) grain content among 225 pearl millets from Sudan, illustrating good scope for biofortification breeding. Significant genetic variation regarding phosphorus efficiency related traits was found among 46 landraces and 54 improved open pollinated varieties under phosphorus-fertilized and phosphorus deficiency conditions (Gemenet et al., Reference Gemenet, Hash, Sanogo, Sy, Zangre, Leiser and Haussmann2015b). Large variability for numerous agro-morphological traits was also reported for pearl millet by Gemenet et al. (Reference Gemenet, Hash, Sy, Zangre, Sanogo, Leiser, Parzies and Haussmann2014, Reference Gemenet, Beggi, Hash, Sy, Sanogo, Zangre, Falalou, Buerkert and Haussmann2015a), Haussmann et al. (Reference Haussmann, Boureima, Kassari, Moumouni and Boubacar2007), Pucher et al. (Reference Pucher, Høgh-Jensen, Gondah, Hash and Haussmann2014) and Upadhyaya et al. (Reference Upadhyaya, Yadav, Reddy, Gowda and Singh2011).
All these studies facilitate the utilization of pearl millet genetic resources. However, no information is so far available about agro-morphological variation and adaptation to WCA of the pearl millet reference collection established by the Generation Challenge Program (GCP). Evaluation of GCP reference sets can serve to identify further trait-specific germplasm. Furthermore, development of genomic tools via association of trait variation with genetic differences could render breeding programmes more efficiently (Pattanashetti et al., Reference Pattanashetti, Dwivedi, Vetriventhan, Reddy, Singh and Upadhyaya2016).
Pearl millet production constraints in WCA include low soil fertility, unpredictable drought, and high pressures of downy mildew (Sclerospora graminicola (Sacc.) J. Schroet.) and the parasitic weed Striga hermonthica (Del.) Benth. While some WCA farmers strive to intensify their production using fertilizer, many are unable to afford/access fertilizer, especially women. Therefore, identification of germplasm that can cope with low soil fertility and simultaneously produce stable yields is crucial. Kountche et al. (Reference Kountche, Hash, Dodo, Laoualy, Sanogo, Timbeli, Vigouroux, This, Nijkamp and Haussmann2013) reported the existence of genetic variation for Striga resistance in pearl millet and demonstrated a significant response to recurrent selection in a Striga-resistant genepool. Further screening of genetic resources for Striga resistance is required so that promising materials can be integrated into the genepool to assure longer-term selection gains or adaptation of the genepool to further environments.
The objectives of this study were therefore to:
(1) extend passport data of different WCA accessions within the GCP pearl millet reference collection;
(2) find sources of Striga resistance in pearl millet;
(3) determine relationship patterns of pearl millet accessions in WCA.
Materials and methods
Selection of genetic material
A total of 81 pearl millet accessions from the GCP pearl millet reference collection, maintained at ICRISAT-India were selected and successfully multiplied for this study (online Supplementary Table S1, Supplementary Fig. S1). The accessions were selected based on their WCA (including Sudan) origin due to a clear lack of adaptation and a very poor performance of non-WCA pearl millets in WCA environments (Padilla, Reference Padilla2007). To cover a potentially higher variability, three accessions were added from South Africa. The 81 accessions comprised 78 cultivated landraces and three improved cultivars. The 78 landraces originated from 13 countries (online Supplementary Fig. S1), namely Benin (1 accession), Burkina Faso (8), Cameroon (6), Central Africa (1), Ghana (4), Mali (9), Mauritania (1), Niger (15), Nigeria (14), Senegal (1), South Africa (3), Sudan (10) and Togo (5). The three improved cultivars originated from ICRISAT-Niger (2) and from a joint ICRISAT-Niger/Institut d'Economie Rurale (IER, Mali) breeding programme (1). Wild accessions were discarded from the collection due to the risk of spreading as weed and contaminating valuable elite germplasm, and seed multiplications of the breeders.
Seed multiplication
Around 10 g of seed were obtained per accession from the collection holder. The multiplication of the 81 accessions was done at ICRISAT-Niger in the off-seasons 2008/2009 and February 2009 using manual ‘sibbing’ which simulates the natural out-crossing behaviour of the species. It consists of the following steps: (i) covering all emerging panicles with a parchment pollination bag; (ii) collecting pollen from several panicles of one accession; (iii) mixing the pollen collected from panicles of the same accession; (iv) distributing the pollen mixture onto other panicles of the same accession where only the female part flowers and no pollen is shed yet (protogyny); and (v) harvesting the (crossed) seed exclusively from the pollinated female panicles. This method largely corresponds to random mating and should reduce any possible inbreeding depression that may have occurred during previous regenerations – unless there were severe bottlenecks (i.e. very low number of plants effectively contributing to the next generation) in previous generations. Low effective population size (N e < 30 plants contributing to the next generation) for some accessions in the first off-season (due to germination problems and low seed quantities) was corrected with the second off-season multiplication effort for the majority of entries. It is therefore assumed that the majority of the genetically heterogeneous pearl millet accessions were properly sampled during the seed multiplication. However, it was not possible to produce enough seed per accession to test them in all environments. Considering thus only 74 accessions were used in all trials. The complete set was used at Sadoré in the Striga resistance trial.
General trial conditions
The field trials were conducted at three locations in WCA (online Supplementary Fig. S1) in 2009: Cinzana in Mali (13°15′10″N, 5°57′55″W), Maiduguri in Nigeria (11°50′0″N, 13°9′0″E), and Sadoré in Niger (13°14′0″N, 2°17′0″E). Planting dates differed considerably at the three sites, ranging from June 16 at Sadoré and July 9 in Cinzana to August 3 at Maiduguri (Table 1). The late planting at Maiduguri was due to religious clashes in July, followed by a drought spell. The trial sites differed in monthly rainfall and mean maximum and minimum temperatures (Table 1).
Table 1. Summary of weather data during characterization trials of West and Central African accessions of the GCP pearl millet reference collection

a In brackets: rains that appeared before planting.
Characterization trials under low-input and fertilized conditions in Niger, Nigeria and Mali
For the agro-morphological characterization, the 74 accessions were randomized together with seven checks in a 9 × 9 α lattice design with three replications. Plot size consisted of one row of 4.8 m length and 0.8 m between hills; rows were 0.75 m spaced (4.2 m2/plot). Following location-specific recommendations, seven hills were planted per plot in Maiduguri and Sadoré, while 10 hills were planted per plot in Cinzana. The seven checks included six improved varieties from WCA and one exclusive local variety at every location. Thinning was done to two plants per hill. The soil fertilization treatment levels were handled as separate environments. The trials were planted either side by side in the same field, or in different fields of the same station. Data collection was done in each plot for the following traits: number of hills after emergence (NHE) and number of hills at harvest (NHH), seedling vigour (SVIG; 1–5, 5 being the best), number of hills infested by downy mildew (DM), days to 50% flowering (FLO; Julian days), panicle exertion (cm), plant height (PHT; cm), panicle length (PLE; cm), panicle circumference (CIR; cm), panicle compactness (CMP; 1–3, 3 being the best), number of productive panicles (NPH), panicle yield (PYD; g/m2) and grain yield (GYD; g/m2). Stover yield (STYD; g/m2) data were collected for Sadoré and Maiduguri, but not recorded in Cinzana at the low-input trial. Thousand grain weight (TGW) was recorded at Sadoré only. From these data, other traits were computed, such as the percentage of downy mildew infestation (DM %), number of productive tillers/plant (TILL), the average panicle weight (APW; g) and the number of grains per panicle (NGP).
The evaluation of Striga resistance in Niger
The Striga resistance evaluation in an isolated field at Sadoré involved 81 accessions from the GCP reference collection plus 18 checks. The 99 entries were randomized in a fourfold replicated alpha design with 11 incomplete blocks per replication and nine plots per incomplete block. Each experimental unit consisted of one row of 3 m length. Spacing was 1.6 m between rows and 0.5 m between hills within rows (7 hills/plot; 5.6 m2/plot). Thinning was done to two plants per hill. The field was artificially infested with Striga seeds collected near Sadoré on pearl millet in the previous year. Data collection in each plot comprised the following traits: NHE, NHH, SVIG, DM, FLO, PHT, PLE, CMP, NPH and PYD. In addition, the following Striga resistance related traits were assessed: Date of Striga emergence (DES; Julian days), number (NSx) and vigour (VIGx; 1–9, as defined by Haussmann et al. (Reference Haussmann, Hess, Welz and Geiger2000), with nine being the best) of emerged Striga plants, with x = at 66, 86 and 105 days after planting. From these data, more comprehensive resistance measures were computed for each entry, such as Striga severity (SEVx; NSx * VIGx) at the different dates, area under Striga number progress curve (ASNPC) and area under Striga severity progress curve (ASVPC) as described by Haussmann et al. (Reference Haussmann, Hess, Welz and Geiger2000).
Statistical analysis
First, the data were analyzed individually per environment (location × fertilization level) to estimate repeatabilities (single environment heritabilities) and to eliminate outliers for further comparisons, using the following model:
where Y ilmn is the observed phenotype; μ is the grand mean; G i is the effect of genotype i; R l is the effect of replication l; B lm, is the effect of incomplete block m nested in replication l; and E lmn the residual effect of the plot n nested in block m and replication l. The adjusted mean for each entry was calculated by regarding genotypic effects as fixed, while the other factors were treated as random. For estimating repeatabilities, the genotypic effects were regarded as random.
Finally, the data were analysed in a one-step approach. The following mixed model was designed under the assumption of heterogeneous error variance of environments:
where Y ijklmn is the observed phenotype; μ is the grand mean; G i is the effect of genotype i; L j, is the effect of location j; F k is the effect of the fertilizer level k; GL ij is the interaction effect of genotype i and location j; R jkl is the effect of replication l in every location × fertilization combination; B jkm, is the effect of incomplete block m for every location × fertilization combination; and E jklmn, the residual effect of the plot n nested in block m, replication l, and for every location × fertilization combination. Adjusted means were calculated by regarding genotypic and fertilization effects as fixed, while the other factors were treated as random. For estimating variance components, the genotypic effects were regarded as random.
Variance components significance was tested using the likelihood ratio test for model comparison (Stram and Lee, Reference Stram and Lee1994). Broad-sense heritability (H 2) was calculated as:
 $$H^2 = \sigma _{\rm g}^2 \times \left( {\sigma _{\rm g}^2 + \displaystyle{{\sigma _{{\rm gl}}^2} \over L} + \displaystyle{{\sigma _{{\rm gf}}^2} \over F} + \displaystyle{{\sigma _{{\rm glf}}^2} \over {LF}} + \displaystyle{{\sigma _{\rm e}^2} \over {LFR}}} \right)^{ - 1}, $$
$$H^2 = \sigma _{\rm g}^2 \times \left( {\sigma _{\rm g}^2 + \displaystyle{{\sigma _{{\rm gl}}^2} \over L} + \displaystyle{{\sigma _{{\rm gf}}^2} \over F} + \displaystyle{{\sigma _{{\rm glf}}^2} \over {LF}} + \displaystyle{{\sigma _{\rm e}^2} \over {LFR}}} \right)^{ - 1}, $$
where ![]() $\sigma _{\rm g}^2 $ represents the genotypic variance,
$\sigma _{\rm g}^2 $ represents the genotypic variance, ![]() $\sigma _{{\rm gl}}^2 $ the genotype by location interaction variance,
$\sigma _{{\rm gl}}^2 $ the genotype by location interaction variance, ![]() $\sigma _{{\rm gf}}^2 $ the genotype by fertilization interaction variance,
$\sigma _{{\rm gf}}^2 $ the genotype by fertilization interaction variance, ![]() $\sigma _{{\rm glf}}^2 $ the genotype by location by fertilization interaction variance, and
$\sigma _{{\rm glf}}^2 $ the genotype by location by fertilization interaction variance, and ![]() $\sigma _e^2 $ the residual variance. L, F and R are the number of locations, fertilization levels and replications, respectively.
$\sigma _e^2 $ the residual variance. L, F and R are the number of locations, fertilization levels and replications, respectively.
The Pearson coefficients of correlation were calculated to determine relationships among the observed traits, using adjusted means. The respective significance levels were Bonferroni-adjusted.
The Additive Main Effect and Multiplicative Interaction (AMMI) analysis was performed based on lattice-adjusted entry means per location for grain yield, in order to assess patterns of genotype by environment interaction. The principal component analysis (PCA) was based on centred adjusted means across environments of all above-mentioned traits having heritabilities higher than 70% to avoid strong environmental bias on the PCA.
All calculations were performed within the R-environment v. 2.14.2 and v. 3.2.3 (R Development Core Team, 2012). Mixed model analysis was conducted using the ASReml v. 3.0 package for the R-environment (Butler et al., Reference Butler, Cullis, Gilmour and Gogel2009) and the AMMI was calculated using the agricolae package v. 1.2-8 (de Mendiburu, Reference de Mendiburu2016).
Results
Differentiation for agro-morphological traits under low-input and fertilized conditions
Genotypic differences among the tested accessions were significant and repeatability estimates were reasonably high for most phenological, morphological and performance traits. Repeatabilities ranged from 16% for seedling vigour at the Cinzana low-input environment to 98% for flowering time at the Cinzana high-input environment (online Supplementary Table S2). When comparing repeatabilities in the low-input versus fertilized treatments across all traits, no clear tendencies could be observed, except for stover yield, where repeatability estimates tended to be higher under high-input conditions. In Cinzana, repeatabilities were lower for seedling vigour and higher for downy mildew susceptibility than in Maiduguri and Sadoré. The ranges of trait expression at each site were large and significant among the tested accessions (online Supplementary Table S2), indicating the existence of, e.g. extra-early or late flowering accessions, accessions with very long or short panicles, downy mildew resistant and high-yielding accessions. Correlation coefficients between the high- and low-input treatments for the single locations (Cinzana, Maiduguri and Sadoré) were all positive and significant (α = 0.05), except for seedling vigour in Cinzana and downy mildew in Sadoré (online Supplementary Table S3).
The combined analysis across sites and soil fertilization treatments revealed highly significant genetic variance for all traits (Table 2). Broad-sense heritability estimates ranged from 0.39 for seedling vigour to 0.96 for panicle length. Separation of the genotype by environment (G × E) interaction revealed significant genotype by location (G × L) effects for all shown traits, except PLE. Genotype by soil fertilization (G × F) interaction was only significant for stover yield and panicle yield. The threefold interaction (G × L × F) was significant for downy mildew susceptibility, plant height and panicle circumference.
Table 2. Means ± standard error (SE) and ranges of the 74 pearl millet accessions, as well as estimated components of variance (V), for genotypic effects (G), genotype by location interaction (G × L), genotype by soil fertilization interaction (G × F), threefold interaction (G × L × F), their standard errors (SE), and broad-sense heritability estimates (H 2) for various traits, from the combined analysis across three sites and two soil fertilization treatments

*, ** Significant at P = 0.05 and P = 0.01, respectively.
Trait abbreviations: SVIG, seedling vigour; DM, downy mildew infestation; FLO, days to 50% flowering in Julian days; PHT, plant height; STYD, stover yield; PLE, panicle length; CIR, panicle circumference; TILL, number of productive panicles per hill; PYD, panicle yield; APW, average panicle weight; CMP, compactness of panicle; GYD, grain yield; TGW, thousand grain weight; NGP, number of grains per panicle.
Genetic variances explained the largest part of the total variance for all traits except seedling vigour and downy mildew susceptibility. The variance ratio of G to G × L, G to G × F and G to G × L × F for seedling vigour was 1:1.5, 1:0 and 1:0 and for downy mildew susceptibility 1:2.3, 1:0 and 1:0.52. This led to relatively low broad-sense heritabilities for these traits (0.39 for seedling vigour and 0.46 for downy mildew susceptibility). However, the variance ratios were 1:0.33, 1:0 and 1:0.07 for plant height, 1:0.02, 1:0 and 1:0.02 for PLE and 1:0.35, 1:0.09 and 1:0 for grain yield leading to high broad-sense heritabilities of 0.87, 0.96 and 0.74, respectively.
The local checks (G81), which differed among locations, had the highest yield in the combined analysis. Five out of six improved checks (G75, G76, G77, G78 and G79) were performing well in the individual environments and were among the eleven best in terms of grain yield in the combined analysis (online Supplementary Table S4). Individual accession means are made available for each location–soil fertilization combination in online Supplementary Table S5.
Relationships among traits
In the combined analysis, high grain yield was significantly associated with high seedling vigour, large plant height, high stover yield, long panicles, high panicle yield and high average panicle weight, strong panicle compactness, a high number of grains per panicle, and low downy mildew infestation (Table 3). The correlation was non-significant for flowering time, panicle circumference and thousand grain weight. The significance of correlation coefficients differed greatly across single environments. For example, seedling vigour and grain yield were not correlated at both Cinzana environments, but highly significantly associated at Maiduguri and Sadoré. High downy mildew infestation, on the other hand, was significantly associated with low grain yield at Cinzana, but the two traits were unrelated at Maiduguri and Sadoré. Flowering time was significantly and positively associated with grain yield only at the high-input environment in Cinzana. Panicle length was significantly associated with grain yield in the combined analysis, at all low-input environments and at the fertilized site at Cinzana. High tillering was associated with higher grain yield at Cinzana and Maiduguri, while there was no significant correlation between these traits in Sadoré.
Table 3. Pearson coefficients of correlation between grain yield and other traits observed for 74 pearl millet accessions at three sites with two soil fertilization levels (high and low), and combined across all environments

*, **, *** Significant at P < 0.05, P < 0.01 and P < 0.001, respectively.
Trait abbreviations: see Table 2.
Patterns of genotype × environment interaction
In the AMMI analysis, principal component (PC) 1 explained 56.9% and PC2 20.2% of the total G × E interaction (Fig. 1). The three locations (Cinzana, Sadoré and Maiduguri) were clearly separated from each other. The high- and low-input sites exhibited similar influence on the genotypes at Cinzana and Maiduguri, with the high-input site showing stronger interaction especially at Maiduguri. However, at Sadoré, high- and low-input environments were clearly separated from each other. The low-input environment was showing little interaction and the high-input environment exerted the strongest interaction of all six environments. Genotypes closer to the centre were more stable over all environments.
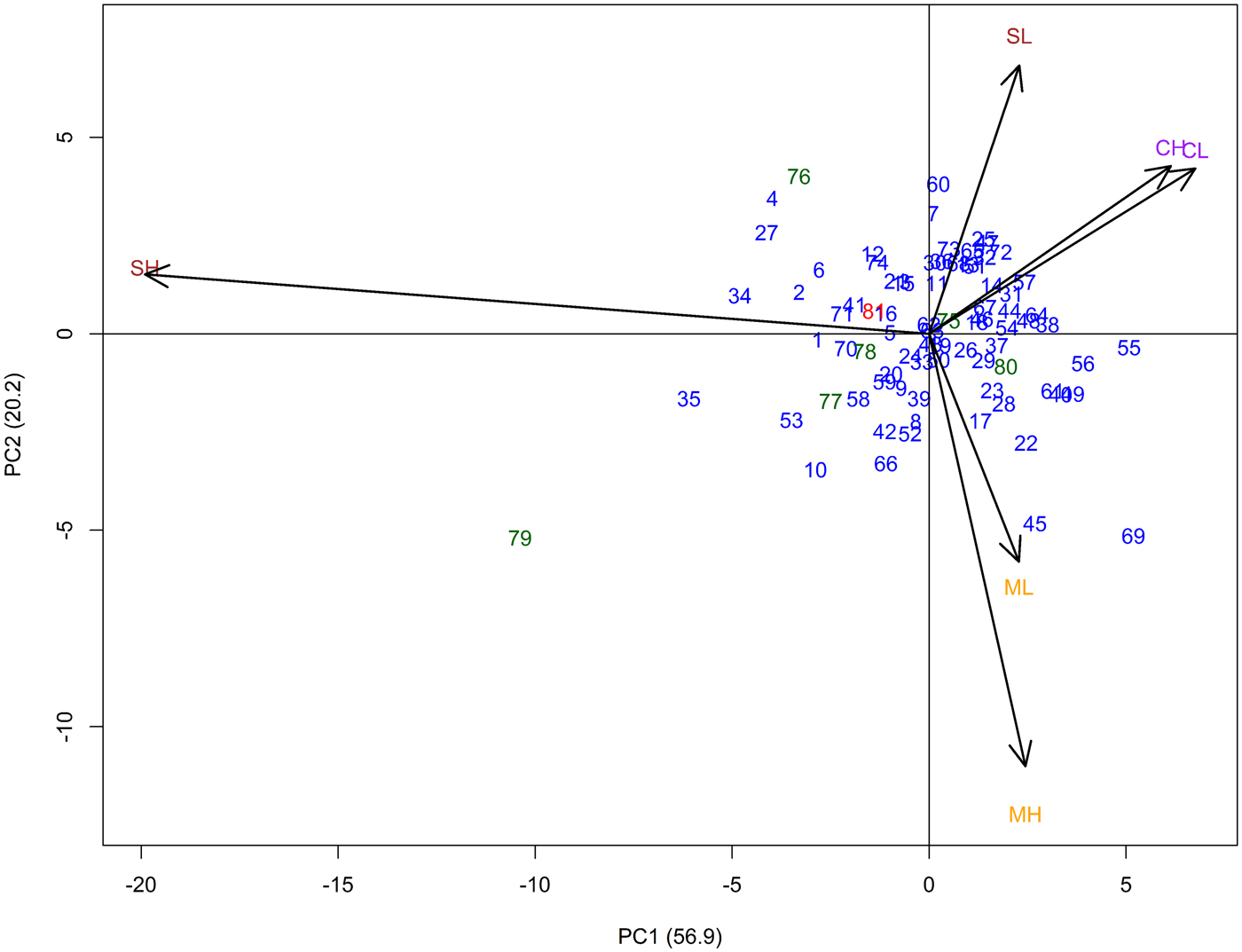
Fig. 1. AMMI biplot illustrating relationships among test sites and entries for 74 GCP pearl millet accessions (blue), six improved checks (green) and the average of the local checks (red) evaluated at three sites (Cinzana (C) in Mali, Maiduguri (M) in Nigeria, and Sadoré (S) in Niger) with two soil fertilization levels per site (low (L) and high (H)-input) using single environment adjusted means (for assignment of entry numbers, see online Supplementary Table S1).
The estimated AMMI stability values (ASV) for grain yield data revealed a wide range of the tested entries (0.2–10.4). Accessions with higher grain yield stability (lower ASV value) compared with the tested checks were identified; however, none of the accessions out-yielded the on average highest-yielding local checks (online Supplementary Table S6).
Diversity of the tested accessions
PC 1 and 2 explained 52.4 and 14.4% of the total variation, respectively (Fig. 2). Accessions could generally not be grouped distinctively by origin. Accessions from Ghana (orange) were negatively associated with all included traits and accessions from Cameroon were among the tallest. However, the observed admixture hinders further association of groups and traits, e.g. accessions from Niger (light-green) were found to be both among the tallest and the shortest (Fig. 2).
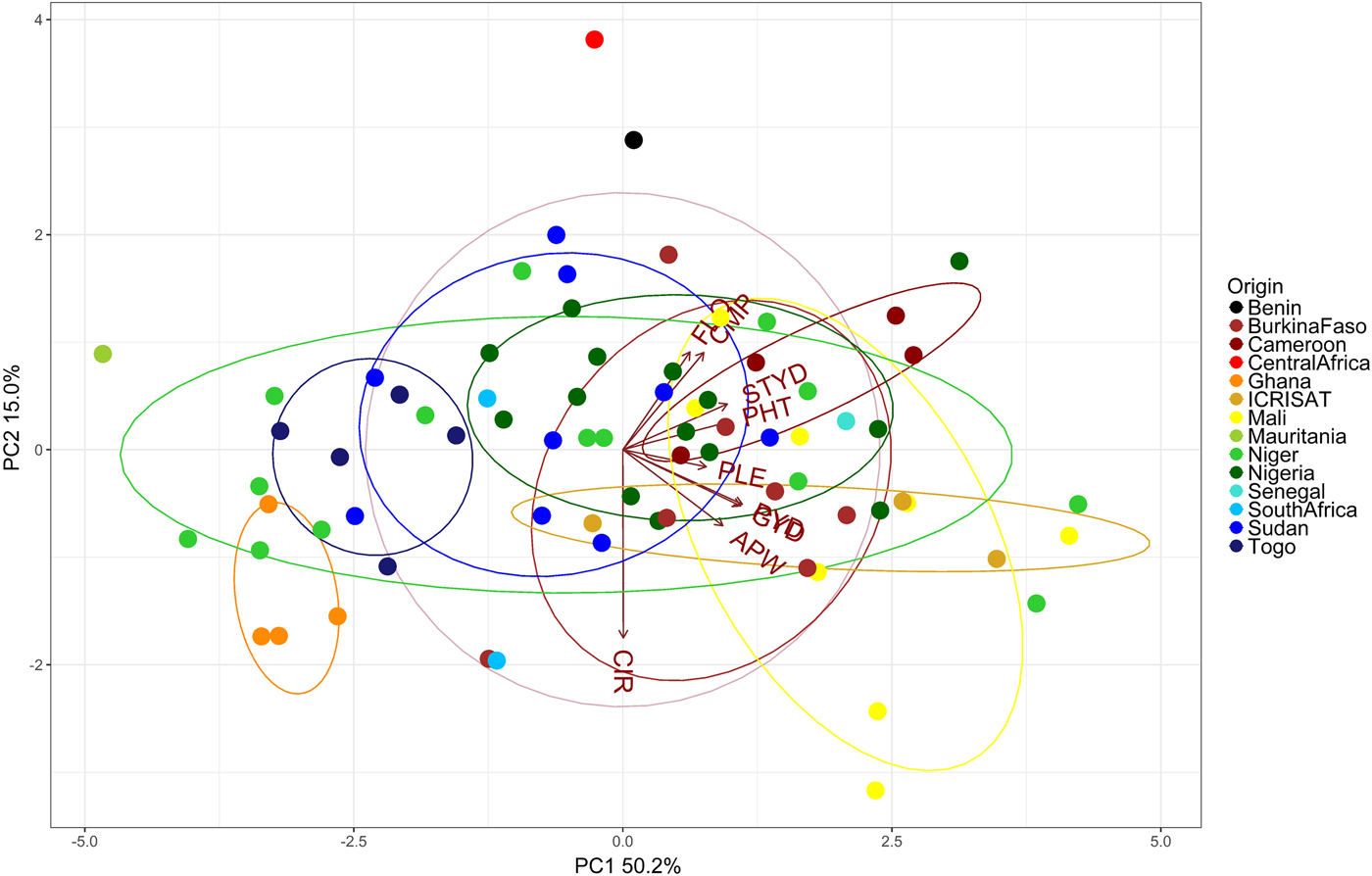
Fig. 2. Principal component analysis (PCA) illustrating relationships between 74 pearl millet accessions (dots) evaluated at three sites (Cinzana in Mali, Maiduguri in Nigeria, Sadoré in Niger) with two soil fertilization levels per site (low- and high-input). Vectors indicate traits (Trait abbreviations, see Table 2).
Evaluation of Striga resistance in Niger
The evaluation of 81 accessions from the GCP pearl millet reference collection for Striga resistance yielded high repeatabilities among the entries for various Striga resistance measures (Table 4). High repeatabilities were also obtained for flowering time (92%) and panicle yield (87%). Certain accessions were found highly resistant to Striga, but none of the accessions out-yielded the check or showed a higher Striga resistance (Table 4). The highest yielding accession was G10 from Mali with 111.2 g/m2 (panicle yield). The accessions G33 from Benin and G64 from Togo were the best accessions regarding Striga resistance (ASVPC). The complete results are shown in online Supplementary Table S7.
Table 4. Repeatabilities (Rep %), accession means ± standard error (SE), ranges, and performance of the best check for various traits estimated in the pearl millet Striga resistance trial conducted at Sadoré in 2009

Trait abbreviations: see Table 2; DES, days to first Striga emerging in the plot in Julian days; NSx, number of emerged Striga plants at x days after planting (dap). SEVx, Striga severity at x dap; MSTR, mean number of emerged Striga across three counts; ASVPC, area under Striga severity progress curve.
Values smaller than zero (due to statistical adjustment) were corrected to zero.
Discussion
General comments on the characterization trials
The three test sites differed largely for planting date, the total amount of rainfall and rainfall distribution. The outstandingly high rainfall amount during September at Cinzana led to the positive association between grain yield and flowering time at this location, and was disfavouring early-flowering accessions whose pollen was thereby washed away. This could be the reason for the moderate yield level at Cinzana despite the high precipitation. The high amount of rain at Cinzana promoted downy mildew development and led to the significant negative correlation between grain yield and downy mildew susceptibility.
The Maiduguri site was characterized by late planting, but the late rains in October, after flowering, helped the plants to produce grains. The differences between the low-input and the fertilized trial at Maiduguri were small for most traits, including grain yield. The high rainfall in the month of sowing (266 mm) caused leaching of the applied fertilizer at Maiduguri. This might have converged the two different fertilization treatments at Maiduguri and led to these similar yield levels.
The Sadoré site received lowest rainfall amounts in September and October, and especially its low-input trial suffered from the combination of low soil fertility and terminal drought stress. As described by Valluru et al. (Reference Valluru, Vadez, Hash and Karanam2010) and Beggi et al. (Reference Beggi, Haussmann, Falalou, Gemenet, Buerkert and Beggi2014), phosphorus is essential for plant development. Drought stress can reduce phosphorus uptake (Hash et al., Reference Hash, Schaffert and Peacock2002; Sinclair and Vadez, Reference Sinclair and Vadez2002). Pearl millet can cope with early drought stress, but is relatively susceptible to terminal drought stress (Mahalakshmi et al., Reference Mahalakshmi, Bidinger and Raju1987). Thus, it can be assumed that in the low-input trial at Sadoré, the plants were not able to compensate the lack of fertilizer and performed much worse than in the high-input trial.
Agro-morphological differentiation among the WCA accessions of the GCP pearl millet reference collection
The present study revealed significant genetic variation among the WCA accessions of the GCP pearl millet reference collection for all agro-morphological traits assessed. The characterization data, made available in online Supplementary Tables S4–S11, can enable breeders all over the world to select trait-specific germplasm for further use in their breeding programmes. The estimated repeatabilities in our study were in general reasonably high for targeting desirable traits in future breeding programs. Also, the estimated repeatabilities per trait were within the same range for both, low-input and fertilized conditions, apart from single exceptions. This indicates that the collection can be useful in breeding programmes targeting intensified conditions as well as breeding programmes targeting low-input conditions. The observed ranges of most agro-morphological traits were of the same magnitude as those reported in similar studies (Bhattacharjee et al., Reference Bhattacharjee, Khairwal, Bramel and Reddy2007; Padilla, Reference Padilla2007; Upadhyaya et al., Reference Upadhyaya, Reddy and Gowda2007, Reference Upadhyaya, Yadav, Reddy, Gowda and Singh2011; Bashir et al., Reference Bashir, Ali, Ali, Melchinger, Parzies and Haussmann2013; Pucher et al., Reference Pucher, Sy, Angarawai, Gondah, Zangre, Ouedraogo, Sanogo, Boureima, Hash and Haussmann2015). The ranges observed in this subset of accessions were, however, smaller than those observed in the complete core and complete mini core collections (Bhattacharjee et al., Reference Bhattacharjee, Khairwal, Bramel and Reddy2007; Upadhyaya et al., Reference Upadhyaya, Yadav, Reddy, Gowda and Singh2011). Two possible explanations are most likely responsible for this deviation. First, the number of evaluated accessions was much smaller in this study than in those mentioned before. Second, the accessions described in those studies were tested in a single environment, while in this study genotypes were tested in six environments, which likewise causes shrinkage of the range towards the mean.
The Striga resistance trial conducted at Sadoré in Niger also revealed highly significant differences among the accessions, and allowed identification of potential new resistance sources. However, these results need further validation, since Striga resistance is usually strongly influenced by genotype by environment interactions (Haussmann et al., Reference Haussmann, Hess, Reddy, Mukuru, Kayentao, Welz and Geiger2001; Badu-Apraku, Reference Badu-Apraku2010; Kountche et al., Reference Kountche, Hash, Dodo, Laoualy, Sanogo, Timbeli, Vigouroux, This, Nijkamp and Haussmann2013). Kountche et al. (Reference Kountche, Hash, Dodo, Laoualy, Sanogo, Timbeli, Vigouroux, This, Nijkamp and Haussmann2013) tested progenies from the Striga-resistant pearl millet genepool in Mali and Niger and found the G × E variance being higher than the genetic variance for Striga resistance, indicating that different varieties ranked best for Striga resistance in Niger versus Mali. Haussmann et al. (Reference Haussmann, Hess, Reddy, Mukuru, Kayentao, Welz and Geiger2001) tested 50 sorghum varieties at two locations in both Mali and Kenya and showed that varieties having high resistance to Striga at one test location were possibly susceptible at another test location. Furthermore, Yoshida et al. (Reference Yoshida, Ishida, Kamal, Ali, Namba and Shirasu2010) used SSR markers to reveal a wide genetic diversity in Striga itself. Therefore, resistant accessions identified in the present Striga resistance trial in Niger should be re-evaluated before being used as resistance source, especially in other countries than our test country Niger.
Trait-specific germplasm identification
The positive correlation between seedling vigour and grain yield in Maiduguri, Sadoré and the combined analysis underlined the importance of quick seedling development for high yields, especially under dryland conditions, which is in accordance to the work of Manga and Yadav (Reference Manga and Yadav1995).
Data from Maiduguri and Sadoré can be used to identify entries with good seedling vigour. Cinzana on the other hand, due to the higher precipitation, was the best site for identification of downy mildew-resistant accessions, and accessions with productive tillering capacity. However, due to usually high genotype by environment interaction for downy mildew resistance (Kountche et al., Reference Kountche, Hash, Dodo, Laoualy, Sanogo, Timbeli, Vigouroux, This, Nijkamp and Haussmann2013), it needs to be validated whether the resistant sources identified at Cinzana in Mali remain resistant and could be used as resistance source also in other sites. The high heritability estimates derived from the combined analysis across locations for flowering time, plant height, panicle length, circumference, compactness and for number of grains per panicle indicated good scope for selection of entries with a specific expression of these traits using means across the three test sites. A precise, trait-specific selection of genotypes is crucial for distinct environmental conditions. Certain traits like downy mildew resistance are essential in more humid regions such as the Sudanean zone of WCA, while early flowering is important to avoid terminal drought stress in the Sahelian zone of WCA (Bidinger et al., Reference Bidinger, Mahalakshimi and Rao1987; Pucher et al., Reference Pucher, Sy, Angarawai, Gondah, Zangre, Ouedraogo, Sanogo, Boureima, Hash and Haussmann2015).
Observed patterns of G × E interaction
Separation of the total G × E variance into interaction variance components with locations (G × L), soil fertilization levels (G × F) and the threefold interaction variance (G × L × F) is recommended to determine the appropriate selection strategy (Annicchiarico, Reference Annicchiarico and Annicchiarico2002). In addition, the size of the genotype by year (G × Y) interaction determines how much priority needs to be given to yield stability over years. In the framework of this study, it was not possible to estimate G × Y interaction. Nevertheless, the genetic variance explained the largest part of the total variance for all traits except seedling vigour and downy mildew susceptibility. By separating the interaction variance for all traits, it could be shown that its largest part was explained by interactions with the location effect. The G × F variance was neglectable and the threefold interaction played only a minor role in this characterization study. In the case of seedling vigour, the large G × L interaction can be explained by the differences in precipitation among the three locations. Kountche et al. (Reference Kountche, Hash, Dodo, Laoualy, Sanogo, Timbeli, Vigouroux, This, Nijkamp and Haussmann2013) and Pucher et al. (Reference Pucher, Sy, Angarawai, Gondah, Zangre, Ouedraogo, Sanogo, Boureima, Hash and Haussmann2015) also found high G × E interactions for reaction to downy mildew. The downy mildew pathogen is, like pearl millet, highly allogamous. The pathogen's virulence can match the resistance of the millet population in regions they occur together (depending on the history of the field), causing the high interaction variance (Pucher et al., Reference Pucher, Sy, Angarawai, Gondah, Zangre, Ouedraogo, Sanogo, Boureima, Hash and Haussmann2015).
In our study, the G × E interaction for grain yield was relatively small compared with results from Bashir et al. (Reference Bashir, Ali, Ali, Melchinger, Parzies and Haussmann2013) and Pucher et al. (Reference Pucher, Sy, Angarawai, Gondah, Zangre, Ouedraogo, Sanogo, Boureima, Hash and Haussmann2015, Reference Pucher, Sy, Sanogo, Angarawai, Zangre, Ouedraogo, Boureima, Hash and Haussmann2016). Nevertheless, G × L interaction was still explaining a significant part of the total variation. The AMMI biplot clearly separated the three locations, thereby emphasizing the importance of local adaption. The positive correlation between grain yield and flowering time at Cinzana underlined the specific adaptation of later flowering genotypes to this location, while grain yield was not correlated with flowering time at Maiduguri and Sadoré.
Low- and high-input fields at Cinzana and Maiduguri were grouped closely together, but Sadoré was depicted differently. The average grain yield performance at Sadore under high-input conditions was outstanding (online Supplementary Table S2). The weather conditions at Sadoré, as described before, did not mask the fertilizer effect (as was the case at the other two locations). This explains the clear separation of the high-input environment from all other trials, actually pointing to the existence of G × F interaction at this location. Gemenet et al. (Reference Gemenet, Hash, Sy, Zangre, Sanogo, Leiser, Parzies and Haussmann2014, Reference Gemenet, Leiser, Beggi, Herrmann, Vadez, Rattunde, Weltzien, Hash, Buerkert and Haussmann2016) also observed specific adaptation to either low soil fertility or intensified conditions among West African pearl millet accessions. These authors also encountered masked fertilizer effects at some locations. Precipitation seemed to determine the location effect and had a high influence on the yield level. While the AMMI analysis identified entries with relatively stable yields across the six test sites, these results need to be treated with caution. Bashir et al. (Reference Bashir, Ali, Ali, Ismail, Parzies and Haussmann2014a) and Gemenet et al. (Reference Gemenet, Hash, Sy, Zangre, Sanogo, Leiser, Parzies and Haussmann2014) tested their pearl millet varieties across several years and observed large G × Y interaction effects. High inter-annual rainfall variability is common in WCA (Haussmann et al., Reference Haussmann, Rattunde, Weltzien-Rattunde, Traoré, vom Brocke and Parzies2012) and contributes largely to these year effects and the G × Y interactions. Multi-year trials are therefore necessary to validate the repeatability of observed G × L and G × F patterns and to identify specific genotypes that suit distinct regions reliably.
Diversity analysis and implications for heterotic grouping
The PCA indicated strong genetic admixture of genotypes, independent of the country of origin, and it was hardly possible to identify distinct clusters. WCA is most likely the centre of origin of pearl millet (Brunken et al., Reference Brunken, de Wet and Harlan1977; Oumar et al., Reference Oumar, Mariac, Pham and Vigouroux2008; Tostain and Marchais, Reference Tostain, Marchais and Serge1993) and a vast gene flow caused by high outcrossing rates of ~85% (Sandmeier, Reference Sandmeier1993; vom Brocke et al., Reference Vom Brocke, Christinck, Weltzien-R, Presterl and Geiger2003) is characteristic for this crop. Drought-tolerant, wind-borne pollen (Hoekstra et al., Reference Hoekstra, Crowe and Crowe2006) might contribute to these high outcrossing rates.
Genetic diversity studies using different molecular markers are likewise concluding that strong admixture among pearl millet varieties is predominant and definition of distinct groups is not evident (Lewis, Reference Lewis2010; Stich et al., Reference Stich, Haussmann, Pasam, Bhosale, Hash, Melchinger and Parzies2010; Bashir et al., Reference Bashir, Ali, Ali, Mohamed, Melchinger, Parzies and Haussmann2014b; Pucher et al., Reference Pucher, Sy, Angarawai, Gondah, Zangre, Ouedraogo, Sanogo, Boureima, Hash and Haussmann2015).
Therefore, genetic diversity studies should be combined with combining ability studies to define initial heterotic groups, which are the foundation for maximum exploitation of heterosis in general and for successful and sustainable hybrid breeding in particular (Melchinger and Gumber, Reference Melchinger, Gumber, Larnkey and Staub1998). The great potential of pearl millet hybrids has already been confirmed (Ouendeba et al., Reference Ouendeba, Ejeta, Nyquist, Hanna and Kumar1993; Bidinger et al., Reference Bidinger, Weltzien, Mahalakshmi, Singh and Rao1994, Reference Bidinger, Raj, Abraha, Ali, Obilana and Jones2005; Yadav et al., Reference Yadav, Weltzien-Rattunde, Bidinger and Mahalakshmi2000) and the initial release of hybrids in India in the mid-1960s sixties contributed largely to the overall productivity. It increased from 5.2 kg/ha/yr (1950–1965) to 24.0 kg/ha/yr (1996–2012) (Yadav and Rai, Reference Yadav and Rai2013). Genetic initial combining ability studies among WCA pearl millets were conducted in pearl millet, but without finding clear patterns (Pucher et al., Reference Pucher, Sy, Sanogo, Angarawai, Zangre, Ouedraogo, Boureima, Hash and Haussmann2016). However, promising hybrids developed from Nigerien and Senegalese varieties were found. More extensive combining ability studies with larger sets of genotypes could help to define heterotic groups.
Implications for conserving pearl millet genetic resources
The local checks (which differed among locations) were among the best entries in terms of mean grain yield at all locations. None of the tested accessions out-yielded the local checks. The generally inferior performance of the gene bank accessions observed in this study and similar results of Padilla (Reference Padilla2007), who found only two accessions out of 504 from a global pearl millet core collection with a superior performance in comparison with local checks, lead to the following hypotheses:
• The accessions conserved in the gene bank outside their country of origin may partially lose their local adaptation over time. This will possibly lead to a lower performance in their original environment. Probable causes are inbreeding depression due to low population size (genetic bottlenecks, for example during cluster bagging) and sampling effects, and natural selection happening in a different regeneration environment.
• Farmer's varieties and improved varieties maintained in a region may change towards better adaptation over time, leading to better performance at the test sites, compared with original seed samples collected a long time ago and stored in a gene bank.
The Food and Agriculture Organization of the United Nations (FAO) commonly recommend conserving seeds ex-situ in gene banks. Nevertheless, the risk of collecting and preserving an undersized number of effective alleles or missing prospective alleles in core collections as well as not having the possibility to adapt the accessions to modern demands or to their changing native environment must not be underestimated (Pistorius, Reference Pistorius1997).
The second hypothesis is supported by the work of Bezançon et al. (Reference Bezançon, Pham, Deu, Vigouroux, Sagnard, Mariac, Kapran, Mamadou, Gérard, Ndjeunga and Chantereau2009), who studied changes in the diversity of pearl millet and sorghum in Niger populations between 1976 and 2003. Concerns that these crops suffer from genetic erosion were not confirmed. However, there was a shift towards earlier flowering at the end of the examined period of 27 years. Brown (Reference Brown1989) suggested core collections of 10% of the complete collection, without exceeding the size of 3000 accessions, to capture ~70% of the total variation. The ICRISAT pearl millet core collection might be too small with 504 out of 21,456 entries (~2.3%) (Upadhyaya et al., Reference Upadhyaya, Reddy and Gowda2007) to cover the large diversity of pearl millet. Especially under the assumption that 5–10% of alleles are lost in every generation, while being maintained and regrown in core collections (Burton, Reference Burton1976). The risk of creating or already having created a ‘bottle neck’ by sampling too little or closely related individual plants should also be taken into consideration.
Conclusions
The evaluation of the WCA pearl millet accessions of the GCP reference collection displayed once more the impressive diversity present in WCA pearl millet. Large significant genetic variation for a multitude of agro-morphological traits was observed. The provided list of adjusted means for numerous traits of all tested genotypes at each site and combined across all sites (online Supplementary Tables S4, S5 and S7) is meant to support NARS (National Agricultural Research Systems) breeders and will facilitate the decision on selecting adequate parents, bearing superior traits, for future breeding programmes.
Within the evaluated subsample, several accessions being well adapted to one of the test environments and/or possessing useful traits were identified. In addition, few accessions with high yield and stability for developing high yielding varieties with a smaller G × E interaction than the local checks were found (online Supplementary Tables S6). These accessions need to be further validated across multiple years. However, none of the accessions was superior to local checks in their respective environment.
The experiences from this project and the mentioned studies underline the importance to develop and test appropriate in situ conservation strategies for plant genetic resources to maintain or enhance their adaptation to changing environments. Secondly, there may be a need to verify present ex situ conservation methods, especially for out-crossing species such as pearl millet, with the following questions to be investigated: Do we maintain sufficiently large population size during collection and regeneration of accessions that represent very heterogeneous and heterozygous population varieties? Can we avoid natural selection effects during regeneration that is performed far away from the geographic origin of the accession? Are there allele frequency changes after a certain number of regeneration events due to sampling effects and (unwanted) selection? If yes, for which traits and at which rate? Nevertheless, the development and preservation of germplasm collections ex situ are important to maintain the rich genetic diversity that exists in pearl millet.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1479262117000272
Acknowledgements
The financial support from the Global Crop Diversity Trust, Trust grant no: GS08024, is gratefully acknowledged. HD Upadhyaya (ICRISAT-India) kindly provided the seed from the ICRISAT genebank in Patancheru, India. In addition, the authors would like to thank the following individuals for technical assistance during seed multiplication and accession characterization: A Abarchi, H Adamaou, D Lankoande, T Boye, A Boubacar (ICRISAT-Niger), S Muhtah (LCRI-Nigeria), M Mariko (IER-Mali). The Ph.D. stipend for FT Sattler was funded by the German Ministry of Economic Cooperation and Development (BMZ, GIZ Project No: 13.1432.7-001.00 and Contract No: 81170266). Finally, we thank the McKnight Foundation Collaborative Crop Research Program for the discretionary research funds provided to BIG Haussmann.








