Introduction
The protist Giardia lamblia (syn. duodenalis, intestinalis) is a worldwide occurring zoonotic parasite that causes dysentery in humans and animals. It has a microaerophilic/anaerobic lifestyle which renders it vulnerable to oxygen and reactive oxygen species (Gillin and Diamond, Reference Gillin and Diamond1981; Lloyd et al. Reference Lloyd, Harris, Maroulis, Biagini, Wadley, Turner and Edwards2000; Paget et al. Reference Paget, Maroulis, Mitchell, Edwards, Jarroll and Lloyd2004). Like other microaerophilic protist parasites, such as Trichomonas vaginalis (Coombs et al. Reference Coombs, Westrop, Suchan, Puzova, Hirt, Embley, Mottram and Müller2004) and Entamoeba histolytica (Arias et al. Reference Arias, Gutierrez, Iglesias and Guerrero2007), G. lamblia possesses antioxidant enzyme pathways for the defence against oxidative stress (Mastronicola et al. Reference Mastronicola, Falabella, Testa, Pucillo, Teixeira, Sarti, Saraiva and Giuffrè2014, Reference Mastronicola, Falabella, Forte, Testa, Sarti and Giuffrè2016). It is well established that the thioredoxin-mediated redox system, comprising thioredoxin reductase (TrxR), the electron shuttle protein thioredoxin, and peroxiredoxins (Lu and Holmgren, Reference Lu and Holmgren2014), plays a central role in the antioxidant defence. Peroxiredoxins reduce harmful hydrogen peroxide to water and molecular oxygen via their catalytic cysteines. To this end, their catalytic cysteines need to be reduced by the catalytic cysteines of thioredoxin first. These, in turn, are reduced by TrxR which harnesses reductive power from NADPH and reduces them via an FAD cofactor (Lu and Holmgren, Reference Lu and Holmgren2014). Moreover, in most organisms, thioredoxin has a large number of protein substrates in addition to peroxiredoxins, including transcription factors and ribonucleotide reductase, rendering it a central redox regulator.
In light of its important function as the reducing enzyme of thioredoxin, TrxR has been repeatedly suggested to have considerable potential as a drug target for antigiardial chemotherapy (Tejman-Yarden et al. Reference Tejman-Yarden, Miyamoto, Leitsch, Santini, Debnath, Gut, McKerrow, Reed and Eckmann2013; Watkins and Eckmann, Reference Watkins and Eckmann2016). Comparably little, however, is known about the components of the thioredoxin-mediated redox system in G. lamblia. Most importantly, no functional thioredoxin, the primary substrate of TrxR, has been identified so far in this parasite and, in fact, no protein with such designation exists in the GenBank or any other database. In contrast, functional thioredoxins have been identified and described in T. vaginalis and E. histolytica (Coombs et al. Reference Coombs, Westrop, Suchan, Puzova, Hirt, Embley, Mottram and Müller2004; Leitsch et al. Reference Leitsch, Kolarich, Wilson, Altmann and Duchêne2007, Reference Leitsch, Kolarich, Binder, Stadlmann, Altmann and Duchêne2009).
In order to fill this gap, we identified candidate thioredoxins in the G. lamblia genome, expressed them in Escherichia coli and tested them for activity with G. lamblia TrxR. In the search of a functional thioredoxin and other interaction/binding partners of TrxR, we also performed co-immunoprecipitation (co-IP) experiments with haemagglutinin (HA)-tagged TrxR in G. lamblia cell extracts. Immunofluorescence microscopy was conducted in order to determine the intracellular localization of the TrxR and peroxiredoxin 1.
Materials and methods
Cell culture
Giardia lamblia WB C6 (ATCC 50803) trophozoites were axenically cultivated in Keister's modified Diamond's medium (Keister, Reference Keister1983) in Nunclon Delta tubes (Nunc, Roskilde, Denmark). The cultures were subcultured every third day. Constituents of the growth medium were purchased from Merck [peptone from casein, yeast extract, sodium chloride, glucose, ammonium iron (III) citrate]. Fetal calf serum was purchased from Biochrom (Bioswisstec AG, Schaffhausen, Switzerland). When large numbers of cells were needed for co-IP, T500 triple layer culture flasks with closed caps (Thermo Fisher Scientific, Waltham, Massachusetts) were inoculated with 60 mL of densely grown G. lamblia culture (equalling six culture tubes), filled to the top with growth medium, and incubated for 48 h. Trichomonas vaginalis G3 cells were cultivated in TYM medium as described (Leitsch et al. Reference Leitsch, Kolarich, Binder, Stadlmann, Altmann and Duchêne2009).
Recombinant expression of thioredoxin candidate proteins in E. coli
Of the three proteins identified as candidates (Table 1) for a functional thioredoxin (GL50803_104250, GL50803_3910, and GL50803_9355), two (GL50803_104250 and GL50803_3910) are almost identical with only two amino acid substitutions (104250 → 3910: Y59C, V113A) in the polypeptide sequence. In order to amplify each of the alleles, the respective genes were first amplified including parts of the upstream and downstream sequences where more differences in sequence could be found. The PCR products obtained with the respective primer pairs (given in Table S1) were cloned into the TOPO pCR™ 2·1 vector (Invitrogen). Subsequently, the coding sequences within the cloned fragments were amplified using appropriate primers (Table S1) and cloned into expression vector pET-17b (Leitsch et al. Reference Leitsch, Burgess, Dunn, Krauer, Tan, Duchêne, Upcroft, Eckmann and Upcroft2011). The reverse primers encoded a 6 × His-tag (C-terminal) for isolation of the recombinant proteins in Ni-NTA columns. GL50803_9355 was amplified by PCR (Table S1) and cloned into a pET151 vector, adding a 6 × His-tag to the N-terminus of the protein. Giardia lamblia peroxiredoxin 1 was also cloned into the pET-17b vector (Table S1). All expression plasmids were transformed into E. coli BL21 (DE3) cells and protein expression and purification was performed according to established protocols (Leitsch et al. Reference Leitsch, Kolarich, Wilson, Altmann and Duchêne2007; Müller et al. Reference Müller, Schildknecht and Müller2013). Recombinant G. lamblia TrxR, recombinant T. vaginalis TrxR and recombinant T. vaginalis thioredoxin were expressed as described (Leitsch et al. Reference Leitsch, Kolarich, Binder, Stadlmann, Altmann and Duchêne2009, Reference Leitsch, Burgess, Dunn, Krauer, Tan, Duchêne, Upcroft, Eckmann and Upcroft2011).
Table 1. The putative thioredoxins recombinantly expressed and studied

TrxR assays in cell extracts and/or with purified recombinant proteins
TrxR assays were performed as described before (Leitsch et al. Reference Leitsch, Kolarich, Wilson, Altmann and Duchêne2007, Reference Leitsch, Kolarich, Binder, Stadlmann, Altmann and Duchêne2009, Reference Leitsch, Drinić, Kolarich and Duchêne2012a , Reference Leitsch, Schlosser, Burgess and Duchêne b ). Reduction of thioredoxin was measured as the readout of DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)] reduction at λ = 412 nm (Δε412 = 13·6 mm-1 cm-1) in a Lambda 25 UV-Vis spectrometer (Perkin Elmer, Waltham, Massachusetts) at 37 °C. Heating of the sample was achieved using a PTP A Peltier temperature programmer (Perkin Elmer, Waltham, Massachusetts). For reduction of DTNB, thioredoxins have to be reduced previously by TrxR. Thus, reaction mixtures contained 2 µg mL−1 G. lamblia TrxR, 40 µg mL−1 T. vaginalis thioredoxin or G. lamblia thioredoxin candidate proteins, 0·5 mm NADPH and 1 mm DTNB. Reactions were buffered in 100 mm Tris, pH 7·5. When reduction of candidate thioredoxins was measured with cell extracts, the same assay mixture was used, but recombinant TrxR was replaced with 50 µg protein mL−1 G. lamblia WB C6 cell extracts. As a positive control, TrxR assays were also performed with T. vaginalis TrxR and T. vaginalis thioredoxin (Leitsch et al. Reference Leitsch, Kolarich, Binder, Stadlmann, Altmann and Duchêne2009), following the same protocol. Reduction of T. vaginalis thioredoxin was also measured with T. vaginalis G3 cell extracts (50 µg protein mL−1).
For the measurement of peroxiredoxin 1 reduction by G. lamblia TrxR, 5 µg mL−1 TrxR and 10 µg mL−1 recombinant peroxiredoxin 1 were added to a reaction buffer similar to the one described above, containing, however, a lower concentration of NADPH (0·2 mm) and 4 mm hydrogen peroxide instead of DTNB. Reduction was measured as oxidation of NADPH at λ = 412 (Δε412 = 6·2 mm-1 cm-1).
Construction of HA-tagged expression constructs
In order to obtain a G. lamblia WB C6 cell line expressing HA tagged TrxR, the trxR gene (GL50803_9827) was fused to the sequence for a 3-fold HA-tag at the C-terminus and cloned into the pPac-VInteg vector (Štefanić et al. Reference Štefanić, Morf, Kulangara, Regös, Sonda, Schraner, Spycher, Wild and Hehl2009). In addition to the gene reading frame, the amplified fragment contained 120 bp of the region 5′ of the start codon in order to express the fused trxR gene under its own promoter. By application of the same procedure, peroxiredoxin 1 (GL50803_14521) and β-giardin (GL50803_4812) were fused to a 3-fold HA-tag. Again, each gene was preceded by its own promoter (100 bp upstream sequence in case of peroxiredoxin, 50 bp in case of β-giardin). The sequences of all primers used are given in Table S1. Transfections of plasmids into WB C6 trophozoites were performed in a BTX Electro cell manipulator 600 (Harvard Apparatus, Holliston, Massachusetts) with the settings 500 V, 800 µF and 720 Ω, and transfectants were selected with puromycin (100 mg L−1) as described (Leitsch et al. Reference Leitsch, Müller and Müller2016).
Co-IP of proteins
Co-IP of proteins bound to TrxR was performed closely following an established protocol (Rout et al. Reference Rout, Zumthor, Schraner, Faso and Hehl2016) using immobilized anti-HA antibodies. However, cross-linking of proteins with Lamont's reagent was omitted. For all co-IP experiments approximately 1 × 109 G. lamblia cells were used.
Mass spectrometric identification of proteins and data analysis
Sample preparations for mass spectrometry, mass spectrometric identification of proteins and ensuing data analyses were performed as described recently (Rout et al. Reference Rout, Zumthor, Schraner, Faso and Hehl2016).
Immunofluorescence analysis and microscopy
A polyclonal antiserum was raised in rabbit against the peptide QMFTTTDVENFPS which is part of the G. lamblia TrxR polypeptide sequence (GenicBio Biotech, Shanghai, China). Before immunofluorescence microscopy, antibodies were affinity-purified as described (Hemphill and Gottstein, Reference Hemphill and Gottstein1996) using recombinant G. lamblia TrxR, isolated from E. coli BL21 (DE3) (Leitsch et al. Reference Leitsch, Burgess, Dunn, Krauer, Tan, Duchêne, Upcroft, Eckmann and Upcroft2011), as bait on a nitrocellulose membrane. For visualization of peroxiredoxin and β-giardin, anti-HA antibodies from mouse were used for the detection of HA-tagged peroxiredoxin and β-giardin in the respective transfected cell lines (see above). Samples of G. lamblia WB C6 trophozoites were prepared for immunofluorescence microscopy as described (Skarin et al. Reference Skarin, Ringqvist, Hellman and Svärd2011) by the use of a Nikon Eclipse 80i and the software Openlab (version 5.5.2). As secondary antibody either anti-rabbit serum was used for the detection of TrxR, or anti-mouse serum for the detection of anti-HA antibody. Secondary antibodies had FITC tags.
Results
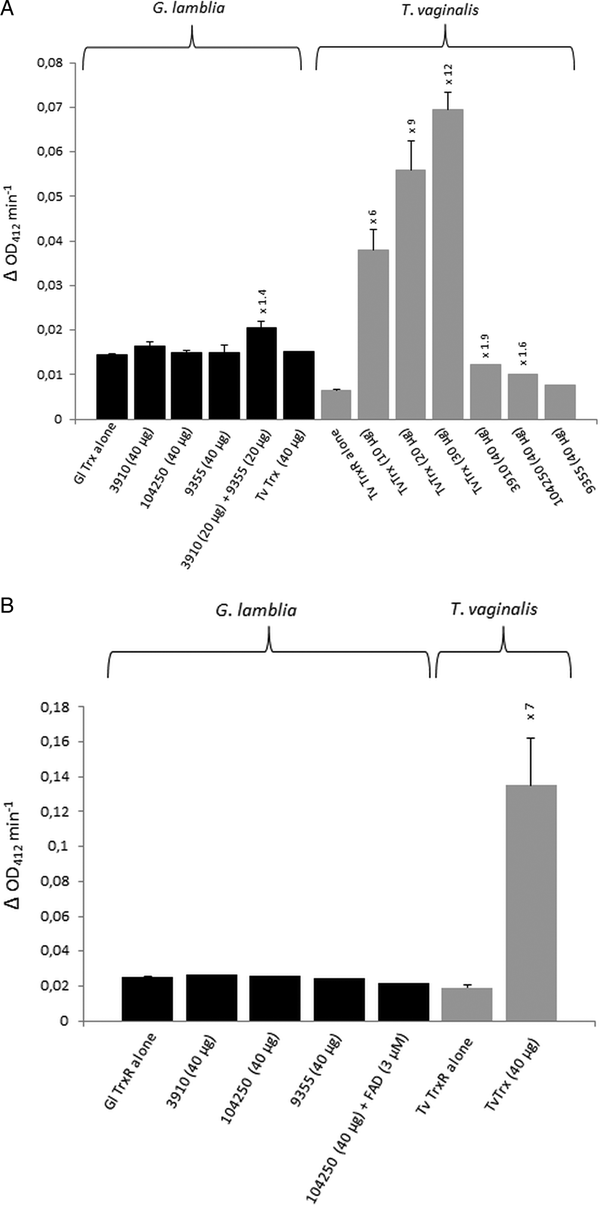
For the identification of potential thioredoxins, we searched the Giardia DB database for proteins that have a thioredoxin-like domain with a CXXC catalytic site and are rather small (10–20 kDa). Apart from protein disulphide isomerase 3 which has a strongly hydrophobic N-terminus and resides in the endoplasmic reticulum (Knodler et al. Reference Knodler, Noiva, Mehta, McCaffery, Aley, Svärd, Nystul, Reiner, Silberman and Gillin1999), three proteins were found to match these criteria: GL50803_104250, GL50803_3910 and GL50803_9355 (Table 1). The first two proteins have a CKDC catalytic site, whereas the third thioredoxin-like protein (GL50803_9355) has a rather unusual CPPC catalytic site and is not closely related to the first two. The three candidate thioredoxins and G. lamblia TrxR were recombinantly expressed in E. coli (Leitsch et al. Reference Leitsch, Burgess, Dunn, Krauer, Tan, Duchêne, Upcroft, Eckmann and Upcroft2011) and used for TrxR enzyme assays. Surprisingly, none of the three proteins was found to be reduced by recombinant G. lamblia TrxR, although the enzyme was functional as indicated by the reduction of DTNB (Fig. 1A). If GL50803_3910 and GL50803_9355 were used in combination, a minimal enhancement of DTNB reduction could be observed (Fig. 1A). Thioredoxin from T. vaginalis (TVAG_125500) was not reduced by G. lamblia TrxR. Further, if NADPH was replaced with NADH reduction of DTNB was also observed albeit at a lower rate (Fig. S1). Importantly, also no reduction of the candidate thioredoxins was observed when G. lamblia WB C6 cell extracts were used instead of recombinant G. lamblia TrxR (Fig. 1B). Addition of FAD, a cofactor of TrxR, did not enhance reduction of DTNB when candidate thioredoxin GL50803_104250 was assayed (Fig. 1B). In contrast, recombinant T. vaginalis TrxR (Fig. 1A) efficiently reduced recombinant T. vaginalis thioredoxin (TVAG_125500) as described before (Coombs et al. Reference Coombs, Westrop, Suchan, Puzova, Hirt, Embley, Mottram and Müller2004; Leitsch et al. Reference Leitsch, Kolarich, Binder, Stadlmann, Altmann and Duchêne2009). Further, strong reduction of T. vaginalis thioredoxin was also observed when recombinant T. vaginalis TrxR was replaced with T. vaginalis G3 cell extracts (205 ± 40 nm min−1 mg−1) (Fig. 1B). Since the measured activity was very similar to the one determined in an earlier study (Leitsch et al. Reference Leitsch, Drinić, Kolarich and Duchêne2012a , Reference Leitsch, Schlosser, Burgess and Duchêne b ), we concluded that our assay was fully functional and, hence, that none of the three candidate G. lamblia thioredoxins tested is a substrate for G. lamblia TrxR. In addition, it was tested if the three candidate G. lamblia thioredoxins could be reduced by T. vaginalis TrxR. Interestingly, a very small enhancement of DTNB reduction could be observed when GL50803_104250 and GL50803_3910 were used, possibly due to unspecific reduction of the disulphides in the candidate thioredoxins (Fig. 1A). The effect was quite comparable to the one observed with G. lamblia TrxR and GL50803_3910 and GL50803_9355 used in combination (Fig. 1A), suggesting that the latter is also due to unspecific reduction of disulphides. Finally, we also tested whether G. lamblia TrxR could directly reduce peroxiredoxin 1 by adding hydrogen peroxide to the assay buffer as substrate but, again, no reduction could be observed.
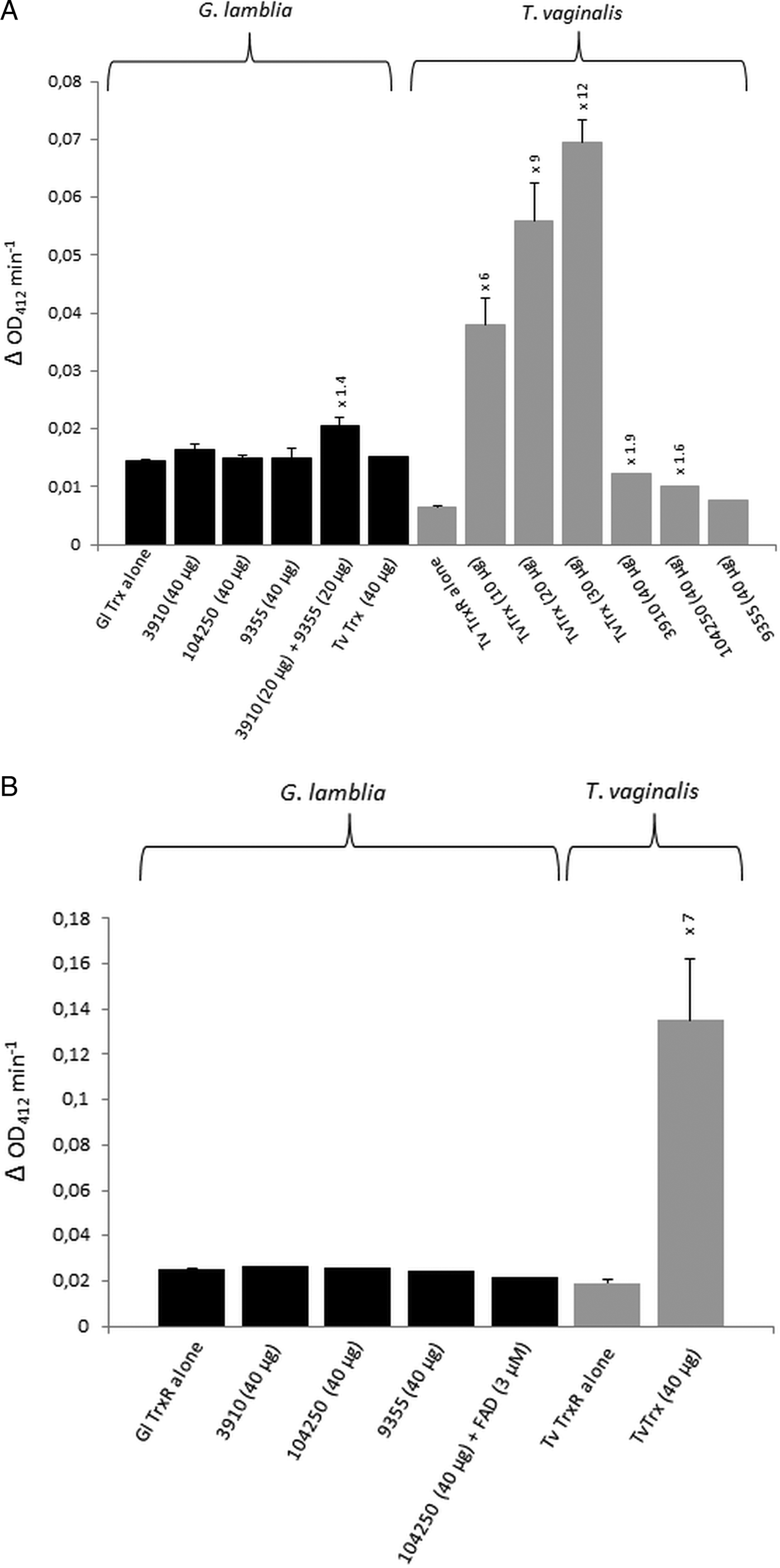
Fig. 1. (A), Reduction of DTNB (at OD412) by thioredoxin reductases (TrxR) of Giardia lamblia and Trichomonas vaginalis either measured in the presence or in the absence of G. lamblia candidate thioredoxins GL50803_3910 (3910), GL50803_104250 (104250) and GL50803_9355 (9355) or T. vaginalis thioredoxin (TvTrx). The amounts of candidate thioredoxins and TvTrx used are indicated. In all reactions, 2 µg mL−1 TrxR were used. The increases in activity of TrxR after addition of a given candidate thioredoxin or TvTrx are indicated above the columns. Measurements with G. lamblia TrxR were performed twice, with the exception of the measurement in the presence of TvTrx, which was only performed once. Measurements with T. vaginalis TrxR were all performed three times with the exception of the measurements in the presence of GL50803_104250 (104250) and GL50803_9355 (9355), which were only performed once. Error bars indicate standard error of the mean (SEM). (B) Reduction of DTNB (at OD412) by cell extracts of G. lamblia and T. vaginalis either measured in the presence or in the absence of G. lamblia candidate thioredoxins GL50803_3910 (3910), GL50803_104250 (104250) and GL50803_9355 (9355) or TvTrx. The amounts of candidate thioredoxins and TvTrx used are indicated. In all reactions, 50 µg protein from extract mL−1 were used. The increase in activity of T. vaginalis TrxR after addition of TvTrx is indicated above the respective column. Measurements with G. lamblia extracts were performed once, with the exception of the measurement in the absence of candidate thioredoxins, which was performed twice. Measurements with T. vaginalis extract were performed three times. Error bars indicate SEM.
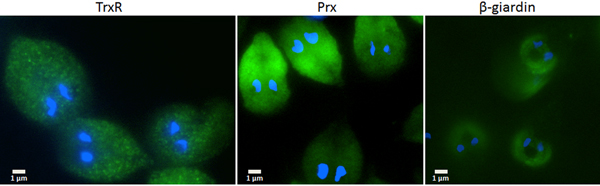
Since the search for a functional thioredoxin had remained unsuccessful, we performed a co-IP experiment for the isolation and identification of protein factors binding to TrxR following an established protocol (Rout et al. Reference Rout, Zumthor, Schraner, Faso and Hehl2016). We argued that a protein with thioredoxin function would likely be isolated together with TrxR. To this end, we constructed a G. lamblia WB C6 cell line expressing an HA-tagged TrxR (TrxR-HA) from a plasmid (Fig. 2) and isolated TrxR protein complexes from cell extracts with immobilized anti-HA antibodies. Isolated protein complexes were submitted for mass-spectrometric and bioinformatics analyses. The procedure was performed twice on two different days (for all peptides identified see Supplementary Table S2), and only proteins which were found to co-isolate with TrxR-HA in both experiments were considered as positives (Table 2). In total, 28 proteins were confirmed to bind to TrxR-HA, including seven metabolic enzymes, seven ribosomal proteins and four enzymes presumably involved in signalling. However, neither a thioredoxin-like protein nor one of the peroxiredoxins co-isolated twice with TrxR-HA. Peroxiredoxin 1 (GL50803_14521) and thioredoxin-like protein GL50803_104250, however, were each found in one of the experiments. Methionine sulfoxide reductase (GL50803_4946), an enzyme known to depend on reduction by thioredoxin (Lu and Holmgren et al. Reference Lu and Holmgren2014), was also found once. As thioredoxin-like protein GL50803_104250 had been found not to be a substrate of TrxR before (Table 1), we performed another set of co-IP experiments with HA-tagged peroxiredoxin 1 (Prx-HA) (Fig. 2). Since peroxiredoxins depend on reduction by thioredoxins in order to be functional (Mastronicola et al. Reference Mastronicola, Falabella, Forte, Testa, Sarti and Giuffrè2016), we hypothesized that a functional thioredoxin might be co-isolated with Prx-HA. As previously, only those proteins were considered as binding partners of peroxiredoxin 1 which were co-isolated twice (Supplementary Table S2). A considerably lower number of proteins, i.e. 10, co-precipitated with Prx-HA as compared with TrxR-HA (Table 3), including three metabolic enzymes, HSP 90-α (GL50803_98054) and a putative ATP-dependent p47 RNA helicase. The only protein which was co-isolated with both baits, i.e. TrxR-HA and Prx-HA, was alcohol dehydrogenase (GL50803_93358), a strongly expressed enzyme in G. lamblia (Leitsch et al. Reference Leitsch, Schlosser, Burgess and Duchêne2012b ). Otherwise, the identified protein sets were different. Thus, the co-IP experiments gave no indication of any firm binding of TrxR or peroxiredoxin 1 to a thioredoxin or other factors commonly assigned to the thioredoxin-mediated redox system, e.g. methionine sulfoxide reductase. Given the conspicuous discrepancy with regard to the proteins co-isolated with TrxR-HA and Prx-HA, we performed immunofluorescence microscopy on WB C6 trophozoites with anti-TrxR and anti HA-antibodies in order to check whether TrxR and peroxiredoxin 1 do indeed localize to the same cell compartment. This was found to be the case as TrxR and peroxiredoxin were both found to localize to the cytoplasm (Fig. 3).

Fig. 2. Schematic presentation of the HA-tagged gene constructs used for co-IP, depicting upstream regulatory sequences (URS), the genes and the 3-fold haemagglutinin tags (3 × HA).
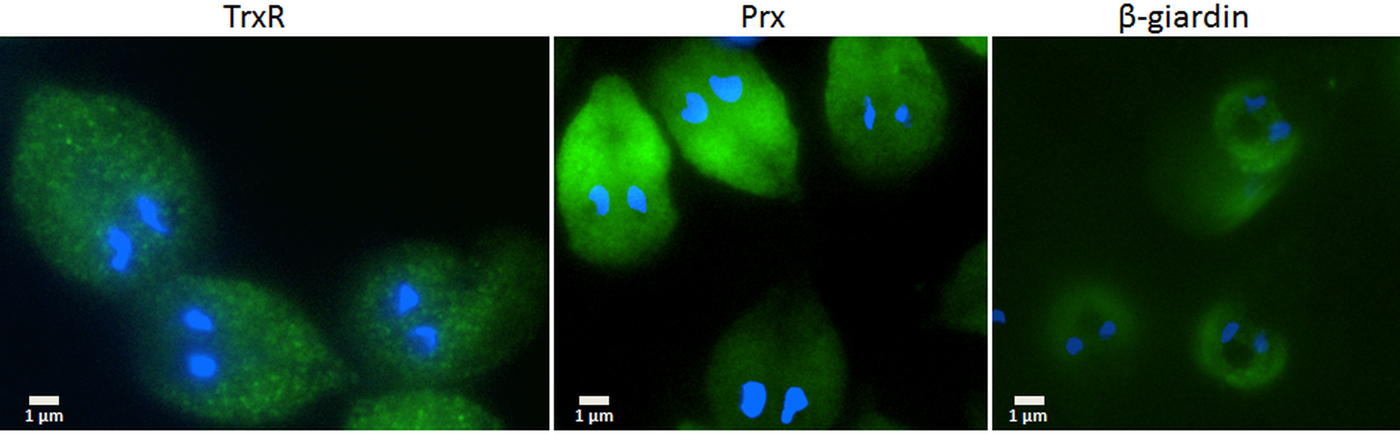
Fig. 3. Immunofluorescent microscopy images of G. lamblia WB C6 cells expressing TrxR (left panel), HA-tagged peroxiredoxin 1 (central panel) and HA-tagged β-giardin (right panel). Secondary antibodies had FITC tags and nuclei were stained with DAPI. TrxR and peroxiredoxin localize to the cytoplasm. β-Giardin, which localizes to the ventral disc, served as a control for the validity of the assay.
Table 2. Proteins isolated with TrxR-HA

Table 3. Proteins isolated with Prx-HA

Discussion
We aimed at the identification of functional thioredoxins in G. lamblia. However, the thioredoxin-like proteins tested were not functional (Fig. 1A and B) and the co-IP experiments conducted with TrxR-HA and Prx-HA did not lead to the identification of any further thioredoxin candidates. Thus, at present our understanding of the thioredoxin system in G. lamblia remains incomplete as there is no evidence for any protein that shuttles electrons from TrxR to peroxiredoxin or any other protein. It was clearly demonstrated that G. lamblia TrxR exerts disulphide reductase activity (Tejman-Yarden et al. Reference Tejman-Yarden, Miyamoto, Leitsch, Santini, Debnath, Gut, McKerrow, Reed and Eckmann2013; Leitsch et al. Reference Leitsch, Müller and Müller2016) but whether it functions as a true TrxR remains unclear. Interestingly, the impact of TrxR activity seems to be rather limited because strong overexpression of the enzyme had only little effect on the viability or physiology of G. lamblia (Leitsch et al. Reference Leitsch, Müller and Müller2016; Müller et al. in preparation). Moreover, the severalfold overexpression as compared to wild-type TrxR of a mutated TrxR which lacks a functional catalytic site had no negative impact on viability (Leitsch et al. Reference Leitsch, Müller and Müller2016). This is surprising, as it would have been expected that the mutated TrxR sequestered factors depending on reduction by TrxR, causing a pleiotropic phenotype. Therefore, it is possible that G. lamblia TrxR only functions as a disulphide reductase but is not involved in any essential protein interactions. The list of proteins co-isolated with TrxR rather supports this notion as none of the proteins, with the exception of enolase (Lemaire et al. Reference Lemaire, Guillon, Le Maréchal, Keryer, Miginiac-Maslow and Decottignies2004), have been described to be associated with the thioredoxin system. Some of the isolated proteins are known to be very highly expressed, e.g. ornithine carbamoyltransferase (unpublished data) and alcohol dehydrogenase (Leitsch et al. Reference Leitsch, Drinić, Kolarich and Duchêne2012a , Reference Leitsch, Schlosser, Burgess and Duchêne b ), or can at least be expected to be highly expressed in the proteome, e.g. HSP70 and ribosomal proteins. This implies that they could interact with TrxR in a rather unspecific manner. Further, the numbers of proteins co-isolated with TrxR-HA differed very strongly between the two experiments (59 vs 541), again suggesting rather unspecific interactions between TrxR and the identified proteins. Arguably, these occur via the reactive cysteine residues of TrxR's catalytic site making it impossible to distinguish between true physiological interactions and artefacts obtained through the co-IP procedure. The numbers of proteins co-isolated with Prx-HA varied less strongly (56 vs 120), but only one protein, i.e. alcohol dehydrogenase, was confirmed to bind to TrxR-HA, as well as Prx-HA. This rather argues against any close interaction of TrxR and peroxiredoxin 1 in a protein complex, although both proteins localize to the cytoplasm. Possibly, reduction of peroxiredoxin 1 by TrxR can be brought about via transient interactions only, without any other further organization of the two proteins in a complex. In contrast to this notion, however, we could not observe any reduction of Prx 1 by TrxR in an in vitro enzyme assay. Finally, it is interesting to note that the spatial organization of TrxR in E. histolytica is different where it distinctly localizes to nodules beneath the cytoplasmic membrane (Arias et al. Reference Arias, Gutierrez, Iglesias and Guerrero2007), suggesting a different role for the enzyme in E. histolytica as compared with G. lamblia.
To conclude, there is currently no evidence for a fully developed canonical thioredoxin-mediated redox system in G. lamblia, although this does not fully rule out that a hitherto undiscovered or wrongly annotated protein could function as thioredoxin. Still, the results of this study bring back into focus a recently presented hypothesis (Mastronicola et al. Reference Mastronicola, Giuffrè, Testa, Mura, Forte, Bordi, Pucillo, Fiori and Sarti2011) which ascribes G. lamblia’s confinement to the small intestine to the weakly developed antioxidant defence of this parasite. The high redox buffering capacity of the small intestine (Blau et al. Reference Blau, Rubinstein, Bass, Singaram and Kohen1999), presumably due to high concentrations of cysteine and glutathione (Mastronicola et al. Reference Mastronicola, Giuffrè, Testa, Mura, Forte, Bordi, Pucillo, Fiori and Sarti2011), limits the extent of oxidative stress to which G. lamblia is exposed, possibly rendering a fully developed antioxidant defence redundant.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/pao.2017.16.
Acknowledgements
We are grateful to our colleagues at Giardia DB for their continuous support.
Financial support
David Leitsch was supported by grant J3492 by the Austrian Science Fund (FWF). Norbert Müller was supported by grant 31003A_163230 of the Swiss National Fund. Samuel Rout was supported by grant 31-140803/1, awarded to Adrian Hehl by the Swiss National Science Fund. Britta Lundström-Stadelmann was supported by grants 31003A_141039/1 and 31003A_160108/1 of the Swiss National Fund.
Conflict of interest
None.