Introduction
After nearly 20 years of collaborative effort involving more than 150 members and their students, and thanks to John Alroy’s long commitment to technical upkeep and scientific rigor, the PBDB stands as one of the most scientifically productive geoinformatics initiatives in the sample-based Earth sciences. To date, the PBDB has enabled more than 235 official publications on such topics as paleobiogeography and latitudinal diversity gradients (e.g., Alroy Reference Alroy2010a; Heim and Peters Reference Heim and Peters2011a; Foote Reference Foote2014; Zaffos and Miller Reference Zaffos and Miller2015; Powell et al. Reference Powell, Moore and Smith2015), taphonomy and the fidelity of the fossil record (e.g., Kowalewski et al. Reference Kowalewski, Kiessling, Aberhan, Fursich, Scarponi, Barbour Wood and Hoffmeister2006; Tomasovych et al. Reference Tomasovych, Fursich and Wilmsen2006; Kosnik et al. Reference Kosnik, Behrensmeyer, Fursich, Gastaldo, Kidwell, Kowalewski, Plotnick, Rogers, Wagner and Alroy2011; Hendy Reference Hendy2011; Heim and Peters Reference Heim and Peters2011b; Smith et al. Reference Smith, Lloyd and McGowan2012; Butler et al. Reference Butler, Benson and Barrett2013), causes and consequences of changes in taxonomic diversity and rates of extinction (e.g., Alroy Reference Alroy2008, Reference Alroy2010b; Alroy et al. Reference Alroy, Aberhan, Bottjer, Foote, Fuersich, Harries, Hendy, Holland, Ivany, Kiessling, Kosnik, Marshall, McGowan, Miller, Olszewski, Patzkowsky, Peters, Villier, Wagner, Bonuso, Borkow, Brenneis, Clapham, Fall, Ferguson, Hanson, Krug, Layou, Leckey, Nuernberg, Powers, Sessa, Simpson, Tomasovych and Visaggi2008, Foote Reference Foote2006; Finnegan et al. Reference Finnegan, Heim, Peters and Fischer2012; Marcot Reference Marcot2014; Darroch and Wagner Reference Darroch and Wagner2015; Kiessling and Kocis Reference Kiessling and Kocsis2015), and paleoecological and morphological evolution (e.g., Hopkins et al. Reference Hopkins, Simpson and Kiessling2014; Foster and Twitchett Reference Foster and Twitchett2014; Klompmaker and Kelley Reference Klompmaker and Kelley2015; Heim et al. Reference Heim, Knope, Schaal, Wang and Payne2015). The impact of the PBDB extends beyond paleobiology to include, for example, constraints on paleogeographic reconstructions (Wright et al. Reference Wright, Zahirovic, Müller and Seton2013) and computer science research involving machine reading and knowledge base creation (Uhen et al. Reference Uhen, Barnosky, Bills, Blois, Carrasco, Carrano, Erickson, Eronen, Fortelius, Graham, Grimm, O’Leary, Mast, Piel, Polly and Säilä2013; Peters et al. Reference Peters, Zhang, Livny and Ré.2014).
Here, we briefly describe the PBDB data model and the Application Programming Interface (API), which enables users to develop custom, independently managed web, mobile, and desktop applications that leverage public PBDB data in near real-time.
The PBDB Data Model
All PBDB records are attributed to references and contributors and belong to one of two components: occurrences and taxonomy (Fig. 1). Although both are widely used and generally understood, there has been up to now little explicit documentation for how PBDB data are managed and combined to produce a result set. In order to properly harness the API, it is important to understand the basics of the data model, outlined below.

Figure 1 Generalized, abbreviated schematic of the PBDB database schema. Boxes labeled “Refs” refer to a single table of references, to which individual data records are linked (records are also linked to contributors). Field names ending in “_no” indicate unique identifiers, stored internally as integers, but should be treated as strings externally. The API can accept for these identifiers integers or a string form that includes support for the Life Science Identifier specification (e.g., Dinosauria can be referred to as either 52775, txn:52775, or urn:lsid:paleobiodb.org:txn:52775). Lookup tables are computed nightly from primary data tables to improve performance of API calls. Many relational tables are omitted for clarity.
Occurrences
PBDB occurrences are taxonomically identified fossils that are grouped into geographically explicit collections (as of August 2015, there were over 172,000 collections containing more than 1.26 million occurrences). The concept of a collection is somewhat nebulous because they vary in scope and purpose, ranging from ecologically-oriented bulk samples from single beds to formation-scale, taxonomically-focused surveys. More than 63% of the collections in the PBDB have a stratigraphic scale that is explicitly stated to be a bed or group of beds, and approximately 75% of all collections are explicitly stated to be outcrop or finer in spatial scale. Some 33% of collections have abundance estimates for their constituent occurrences and approximately 25% have museum repository information and/or specimen numbers. Lithostratigraphic information (e.g., formation) is given for more than 75% of collections and many of the remainder are from deposits lacking such nomenclature; almost 80% have basic sedimentological descriptors. Additional information is also accommodated, ranging from taphonomic attributes and specimen-based size measurements, to collection methods.
All collections are linked to one or more separately managed chronostratigraphic intervals. Age assignments are currently static unless manually edited. All PBDB collections are assigned paleogeographic coordinates based on their present-day latitude/longitude and geologic age using rotation models provided by Christopher Scotese and the GPlates API (http://www.gplates.org).
Each PBDB occurrence has a taxonomic name and, optionally, modifiers expressing the confidence in and resolution of that name (aff., cf., ex gr., sensu lato, ?, “informal”). However, occurrences contain no direct systematic information. Instead, classification is inherited dynamically, as described below.
Taxonomy
The taxonomic apparatus of the PBDB is a stand-alone resource designed to account for the multiple, often conflicting, opinions that exist in the literature. There are two components: authorities, or taxonomic names from an authoritative reference (which may or may not be the reference that originally named the taxon), and opinions, which express the status of and relationships among those names (as of August 2015, there were more than 573,000 opinions on nearly 327,000 authorities). Because only opinions on relationships between names, not their ranks, are used to generate the tree, both Linnean ranked names and unranked clade names are accommodated. Opinions vary in their basis and are assigned any one of the following ordinal values: “stated with evidence”, “stated without evidence”, “implied”, and “second hand”. Authorities and opinions are combined to generate a working taxonomy using a multi-step algorithm, summarized as follows:
1. Opinions are first ranked by their basis (“stated with evidence” taking highest rank) and then by recency of publication.
2. Names with opinions explicitly identifying them as variants of each other (i.e., recombinations, rank changes, and variant spellings) are grouped together. From among all opinions for the members of each group, the highest-ranked opinion (from step 1) is selected as the “classification opinion.” If multiple spellings occur, a variant not marked as a misspelling is selected. Each group is then treated as a unitary name, identified by the original (earliest published) variant (orig_no).
3. Names for which the classification opinion expresses synonymy are then grouped together with their senior synonym, defined as a taxon with a classification opinion explicitly identifying it as a child of another taxon. The highest-ranked (from step 1) classification opinion for each synonym group is taken to be authoritative.
4. Synonym groups are then arranged into a hierarchy according to the classification opinion on each senior synonym (parent_no). Opinions either place them as valid names belonging to other taxa or as invalid names belonging to other taxa (i.e., nomen nudum, nomen dubium, nomen vanum, nomen oblitum, “invalid subgroup of”, “misspelling of”).
5. Each name is then associated with an “accepted name” (accepted_no). For junior synonyms, this is the senior synonym. For invalid names, it is the parent taxon. All other names are their own accepted name. Any chains are then collapsed, so that the accepted name will always be a valid name that is not a junior synonym.
6. The hierarchy is then traversed to compute secondary attributes (e.g., first and last occurrences, number of occurrences, ecological properties, common names, etc.) for each taxon based on the attributes of all subtaxa and supertaxa.
Because of this procedure, there is no taxonomy applied by fiat in the PBDB. Users can influence the impact that an individual reference has on the dynamically generated taxonomy by changing the stated basis of its opinions (e.g., from ‘stated with evidence’, to ‘stated without evidence’, thereby down-ranking the reference’s opinions), but the system aims to be an objective and principled reflection of the literature that it represents. Perl code implementing the taxonomic algorithm described above is accessible at https://github.com/paleobiodb/pbdb-new.
Taxonomy of Occurrences
PBDB occurrences have up to three taxonomic designations: (1) the taxonomic name by which the occurrence was originally identified (required), (2) the most recent re-identification (if any), and (3) the currently accepted name (dynamically generated, as described above). Occurrences do not acquire a classification until their taxonomic name is linked to an authority record. Thus, it is possible for an occurrence to have a valid species-level name stored as a text string, but only genus-level resolution in the taxonomy. Currently, nearly 40% of all occurrences have species- or subspecies-level authorities, and another approximately 25% of all occurrences have unclassified species names assigned to them (i.e., the species name has not yet been entered as an authority record). Approximately 89% of all occurrences have a genus or finer authority record.
Taxonomic data entered into the PBDB automatically propagates to all relevant occurrences and newly entered occurrences automatically acquire relevant taxonomic data. The taxonomy is, of course, only as good as the underlying data that it draws upon. The remedy for any perceived deficiencies is entering the relevant taxonomic references or, if the literature does not yet exist, performing a systematic study, publishing it, and then entering the relevant data.
The PBDB Application Programming Interface
General Purpose
APIs provide a set of protocols and tools for building software. In the context of databases, an API is a specification for how to make remote requests for data (via a standard protocol, such as HTTP) using a semantic that does not require any knowledge of the database software and that returns data formatted in ways that are not specific to any one end use. Although there are few widely agreed upon best practices, the PBDB API has many properties of a representational state transfer (REST) system, meaning, among other things, that specific data resources are uniquely identified by uniform resource locators (URLs), for example:
https://paleobiodb.org/data1.2/taxa/list.txt?name=Otarion&rel=all_parents&show=attr
This returns basic classification information for the trilobite genus Otarion, including all of its parent taxa (rel=all_parents) and their authors (show=attr). The same data are accessible on the classic PBDB website under the “Classification” tab:
https://paleobiodb.org/cgi-bin/bridge.pl?a=checkTaxonInfo&taxon_name=otarion
Both the PBDB API and the PBDB website have a base URL address identifying the server (https://paleobiodb.org), a path identifying a general class of data, and parameters (always preceded by “?” and separated by “&”) identifying specific data elements accessible in that path. The same API URL can be used to obtain data for any taxonomic name in the database by replacing the value of the “name” parameter (e.g., list.txt?name=Bovidae). The identification of a data resource is separated from the format in which the information is returned, meaning that all data can be obtained in any of the available formats (i.e., delimited text, JSON).
Although both the API and PBDB website can return the same data, the latter embeds the response within HTML specific to the purpose of rendering in a web browser. The API, by contrast, returns only a set of field names and values. Thus, the same API calls could be used to build many different remotely hosted web pages, each with styling that is tailored to the needs and tastes of its users. The same API calls could also be integrated into R, Matlab, or Python scripts, called from within a mobile application, used to link data in another database, or included in a publication to identify a data set.
Available Operations
Table 1 summarizes the operations that are currently available in the PBDB API. These operations are grouped into categories organized around specific record types; some return records of that type and others return related records. The API is explicitly versioned in the URL (i.e., /data1.2/) to ensure that it behaves as expected when deployed in applications. Future API changes that impact formatting of responses or accepted parameters will be released with a new version number and previous versions will continue to operate as expected. Documentation for the PBDB API, versions, and examples are provided at the root URL (https://paleobiodb.org/data/).
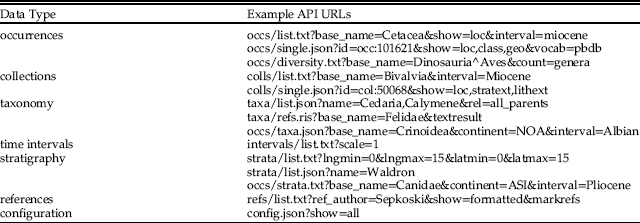
Table 1 Summary of operation types provided by PBDB API version 1.2. The URL prefix for each operation is https://paleobiodb.org/data1.2/. Example operations are given for each data type, including a suffix that specifies the format of the returned data (i.e., “txt”, “json”, “ris”), and a question mark followed by parameters that identify specific data of interest. See online documentation for additional examples and documentation of all parameters and responses.

API Usage Examples
Until recently, the only way to obtain data from the PBDB was via user interaction with a web form (https://paleobiodb.org/cgi-bin/bridge.pl?a=displayDownloadForm). When properly completed and submitted, the form prompts the server to retrieve a defined set of data, process them, and generate a delimited text file, which the user is then prompted to download. Configuring (and understanding) the hundreds of options on the classic PBDB download form takes some effort, and the process must be repeated each time a new data set is desired. At the simplest of levels, the PBDB API can be thought of as a way of specifying options in the PBDB download form and then saving those options for later use as a URL. For example, if one were interested in the present-day and paleogeographic coordinates of Mesozoic echinoderm occurrences, excluding crinoids, along with their original identifications, current traditional Linnean classifications and geological descriptors, the appropriate fields could be completed on the PBDB download form or the following API URL could be used:
Generating a properly formatted API call, like a properly completed download form, takes some effort (i.e., reading the documentation at https://paleobiodb.org/data1.2/). However, once configured, a URL defines a PBDB data set and it can be used repeatedly.
As another simple “bookmark-type” use case, the PBDB API can be invoked to quickly see what new, publicly accessible data have been entered for a particular taxon (or time interval, or geographic region, or any other aspect of interest). This task would be cumbersome via the classic PBDB download form, but it is easy with the API. The following URL retrieves the four most recently entered, publicly accessible Cetacean and Sirenia occurrences and returns who entered and modified the occurrences along with primary references (the option “vocab=pbdb” makes the field names longer and easier to read visually):
This JSON-formatted response is visible in a standard web browser, but to retrieve the data as delimited text simply replace “list.json” with “list.txt” or “list.csv”. A bookmark could be created for this URL, allowing the user to get one-click updates on all recently entered occurrences of specific interest. Elaborating upon this, one could build an application, such as an iPhone app, that used this same type of API call to obtain customized data of interest, which could then be automatically pushed as a user notification or displayed in a visually appealing, interactive way.
The PBDB API can also be used to generate customized, high-level summaries of database content. For example, the following API call returns basic genus-level diversity metrics, with subgenera elevated to genera, for European, non-Avian dinosaurs, using international stage time bins and defaults applied for handling of imprecisely resolved collections and taxa:
Although most users will want to obtain raw occurrence data and then process them using their own analytical procedures to arrive at a diversity estimate, this API call could be useful for building educational or basic data exploration tools.
Finally, a more complex use case is the PBDB Navigator web application (https://paleobiodb.org/navigator), which obtains all of its data from the API. This means that Navigator could have been built by anyone, not just by the group who happens to have direct control over the PBDB server. This also means that a different application, with a completely different approach to searching for and displaying PBDB data, could be constructed using the same API calls. Other examples of applications that leverage the PBDB API are found on the PBDB Apps page: https://paleobiodb.org/#/apps.
It should be noted that most API data derive from a set of computed lookup tables (Fig. 1) that are engineered to reduce server response time and computational load. Because these tables are currently computed once every 24 hours, any new data or changes to data require a 24-hour cycle before they appear in the API. Future extensions to the API framework will allow data updates and additions to propagate throughout the system in near real time.
API Data Use and PBDB Citation
PBDB contributors continue to have the option of placing time-limited access restrictions on the data they enter so as to enable their use prior to public release, which occurs after one year for literature-based data and after five years for unpublished data. In 2013, the PBDB Executive Committee voted to apply a CC BY 4.0 International License to all publicly released PBDB records. Thus, anyone is free to copy, redistribute, adapt and build upon public PBDB data for any purpose, provided that attribution is given and that any changes to the data are indicated. Full attribution includes acknowledgement of the PBDB, citation of original references, and acknowledgement of PBDB contributors. When used in publications, an official publication number should also be requested (https://paleobiodb.org/#/publications). Users of the API can simply include any URLs and may cite this reference.
Summary
Owing to the dedication of John Alroy, Charles Marshall, Arnie Miller, Matthew Kosnik, and many others, and thanks to an international team of several hundred contributors and their students and postdocs, the PBDB has grown into a paleontological resource with broad utility. The API makes it possible for others to participate in the creative process of leveraging PBDB data by developing their own software applications for visualization and analysis. We hope that this, in turn, will stimulate interest in growing and improving all aspects of the underlying data. Future extensions to the PBDB API will include immediate propagation of new data and edits to computed API lookup tables and the capacity for authorized client software to submit data for validation and entry. The latter will open PBDB development to new data acquisition and curation tools that are tailored to the specific needs of field- and museum-based paleontologists. We hope that this capacity will ultimately help to improve the pace at which new and much needed paleontological field- and museum-based data are generated.
Acknowledgements
We thank J. Alroy, C. Marshall and the entire PBDB team over the past 20 years. We also thank current Executive Committee chair M. Uhen, secretary J. Sessa, and members of the Executive Committee and Advisory Board, past and present, for their service. We also thank S. Holland and two anonymous reviewers for comments and suggestions that improved the clarity of this paper. Development of the PBDB API and Navigator supported by National Science Foundation EAR 0949416 and the University of Wisconsin-Madison Dept. of Geoscience. This is Paleobiology Database Publication 237.