This paper contains supplementary material that can be found online at http://journals.cambridge.org/orx
Introduction
A great deal of effort goes into protecting threatened and/or important species. In some cases several threatened species can be protected under the same programme. Nevertheless, when these threatened species include both predator and prey in the same ecological system there may be potential conflict of management plans. This is illustrated by examples from the Channel Islands off the Californian coast. These islands were previously densely populated by the island fox Urocyon littoralis. The arrival and spread of the golden eagle Aquila chrysaetos to these islands in the 1980s, facilitated by the availability of the previously introduced feral pig Sus scrofa as prey, led to a drastic decline in fox numbers (Roemer et al., Reference Roemer, Donlan and Courchamp2002), to the point that breeding programmes were established to save the fox from extinction. The dilemma was: should the golden eagle, which is a protected species, be removed to save another protected species, the island fox? To mitigate the decline in fox numbers, golden eagles were translocated, and feral pigs eradicated from the islands (Courchamp et al., Reference Courchamp, Chapuis and Pascal2003). A similar case in these Channel Islands is that of the threatened San Clemente loggerhead shrike Lanius ludovicianus mearnsi, which is predated by the island fox; a control programme was established to save the shrike (Roemer & Wayne, Reference Roemer and Wayne2003).
Predator removal is commonly practised by wildlife managers. A recent review suggested that predator removal increased hatching and fledging success and breeding populations of birds (Smith et al., Reference Smith, Pullin, Stewart and Sutherland2010). Predation by wolf Canis lupus, puma Puma concolor and coyote Canis latrans may be a significant mortality factor for medium or large ungulates (e.g. deer, caribou Rangofer tarandus, moose Alces alces) in some areas under certain conditions (reviewed in Ballard et al., Reference Ballard, Lutz, Keegan, Carpenter and De Vos2001). A predator removal programme may not affect prey populations that are near carrying capacity but may significantly increase ungulate populations that are substantially below carrying capacity. Thus results of predator removal practice are confounded by numerous factors and only through intensive studies can predation be identified as a major limiting factor (Ballard et al., Reference Ballard, Lutz, Keegan, Carpenter and De Vos2001). Wasser et al. (Reference Wasser, Keim, Taper and Lele2011) studied the effect of predator removal on caribou populations and concluded that management should prioritize the control of human activities before implementation of more extreme actions such as removal of wolves. In other words, resource selection and its physiological consequences can significantly influence predator–prey interactions, and should be considered in management programmes that aim to manipulate complex ecosystems.
In this study we present a problematic case of conflicting conservation policies involving several threatened species. A small isolated population (20–30 individuals) of the mountain gazelle Gazella gazella acaciae inhabits a 6 km2 acacia forest near Yotvata in the southern Arava Valley, Israel (Yom-Tov & Ilani, Reference Yom-Tov and Ilani1987; Dolev & Perevolotsky, Reference Dolev and Perevolotsky2004). A recent genetic survey of gazelles in the Middle East showed that this isolated population is genetically different from the mountain gazelle Gazella gazella gazella in northern Israel but similar to G. gazella from northern Saudi Arabia (Wronski et al., Reference Wronski, Wacher, Hammond, Winney, Hundertmark and Blacket2010). This Critically Endangered G. g. acaciae population has been in decline since the 1960s, presumably because of low calf survival, low genetic diversity, anthropogenic effects and climate change (Yom-Tov & Ilani, Reference Yom-Tov and Ilani1987; Dolev & Perevolotsky, Reference Dolev and Perevolotsky2004; IUCN, 2008a). The congeneric dorcas gazelle Gazella dorcas is a more common species in the Negev but at a regional scale it is categorized as Vulnerable (Dolev & Perevolotsky, Reference Dolev and Perevolotsky2004; IUCN, 2008b) because of intensive motorized hunting and habitat degradation caused by overgrazing and droughts (Mallon & Kingswood, Reference Mallon and Kingswood2001). The law in Israel protects both gazelle species and poaching in the south of Israel is rare. The main alleged cause for the decline of both the mountain and dorcas gazelles in the Negev is predation by the Arabian wolf Canis lupus arabs and the caracal Caracal caracal, a hypothesis based on anecdotal data (Shalmon, Reference Shalmon2003, Reference Shalmon2006). The mountain and dorcas gazelles differ in their laying-up period, which is longer in the mountain gazelle, suggesting greater vulnerability of mountain gazelle fawns to predation (Mendelssohn et al., Reference Mendelssohn, Yom-Tov and Groves1995; Yom-Tov et al., Reference Yom-Tov, Mendelssohn and Groves1995). In response to the decline in gazelle populations in the southern Negev the Israel Nature and Parks Authority (INPA; the government agency that manages all natural resources in Israel) instigated a wolf removal programme that eliminated 37 individuals from the area during 2005–2008. By mid 2006 the isolated and small population of the mountain gazelle was completely fenced and the wolf removal programme was terminated in early 2008.
Although C. lupus is categorized as Least Concern on the IUCN Red List (Mech & Boitani, Reference Mech and Boitani2010), the Arabian subspecies is threatened regionally and is vulnerable locally (Dolev & Perevolotsky, Reference Dolev and Perevolotsky2004; Sillero-Zubiri et al., Reference Sillero-Zubiri, Hoffmann and Macdonald2004) because of systematic shooting and trapping (Mendelssohn, Reference Mendelssohn1983; Cunningham & Wronski, Reference Cunningham and Wronski2010). In Israel the Arabian wolf is fully protected by law and is not persecuted except for specific management actions. Although Hefner & Geffen (Reference Hefner and Geffen1999) have studied some behavioural aspects of this subspecies, no reliable demographic data are available. Hefner & Geffen (Reference Hefner and Geffen1999) documented the dispersal of individuals over 50–150 km, suggesting a continuous admixture between wolves in the southern and central Negev and the likelihood that the wolves in the Negev Desert comprise a single population.
Israel is divided into four INPA districts that are relatively independent of each other but controlled by a single directing body. The 13,000 km2 Negev Desert, which covers more than half of Israel, is divided into two INPA districts (Fig. 1). During 2005–2008 these two districts had different management policies for wolves. The Eilat district culled wolves by trapping or shooting (B. Shalmon, pers. comm.) and tightly controlled the access of carnivores to rubbish pits, although wolves may still have fed from garbage containers inside settlements and army camps. The South district, on the other hand, did not cull wolves and did not control access of wolves to local rubbish pits. Consequently, while the Eilat district attempted to reduce the wolf population to a minimum, the management exercised by the South district counteracted this. The above actions taken by INPA raise a question: how can a single population be managed by two contradictory policies? To address this issue we studied the genetic population structure and estimated population size using faecal DNA. Specifically, we examined whether the wolf population in the two INPA districts has similar densities and genetic composition. The findings of our study offer a new and reliable baseline for modification of the current INPA management policy for the Arabian wolf in the Negev Desert. Furthermore, our results can shed light on the effects of different management policies on the population genetics of a threatened species.

Fig. 1 Distribution of the 12 feeding sites (black triangles) used for collecting wolf Canis lupus arabs scats in the Negev Desert (see text for details). White circles denote locations where scats were collected. Kernel densities of 90, 50 and 10% for wolf scats are denoted. A single smoothing parameter of h=10 km was used in all kernel contours. The heavy dashed line indicates the boundary between the south and Eilat INPA districts.
Study area
The central and southern Negev is a rocky desert, a mixture of mountains, washes, and depressions. The 160 km Arava Valley, an elongated depression of the Rift Valley, borders the Negev in the east. Elevation in the mountainous regions is 400–1,000 m, falling to 50–150 m at the bottom of the Rift. The climate is hot and arid, with an annual rainfall of 50–100 mm, falling during the winter months of November–March (Evenari et al., Reference Evenari, Shanan and Tadmor1982). There are two cities, Eilat (population 50,000) at the northern tip of the Red Sea, and Mitzpe Ramon (population 6,000) in the Negev highlands. All other settlements are agricultural communities (10 in the Eilat district and eight in the South district), mostly in the Rift Valley.
Methods
Sample collection
To sample wolf scats equally over the central and southern Negev Desert we established a grid of 12 feeding sites c. 20 km apart. Five feeding sites were established in the Eilat district (sampling an area of c. 2,129 km2) and seven in the South district (sampling an area of c. 2,910 km2; Fig. 1). In setting out this grid of feeding sites we also considered vehicle accessibility and other entry restrictions. Mean home range size of Arabian wolves in the Negev Desert is 34.6 ± SD 19.5 km2 and mean daily travel distance is 13.7 ± SD 6.7 km (maximum recorded distance travelled in a day is 42 km; Hefner & Geffen, Reference Hefner and Geffen1999). Considering this mean daily travel distance all wolves in this area could probably reach at least one of the feeding sites on a daily basis. Once a month INPA rangers placed a cow calf carcass at each feeding site and scanned the site for scats 1 week later. Each calf carcass was secured to prevent removal by predators. The meat provided at each feeding site was insufficient to support local wolves but provided an incentive for resident wolves to inspect the site on a regular basis and defecate nearby. Because it is difficult to find wolf scats by random searches, except around garbage dumps, this system facilitated the sampling of individuals that are not associated with human habitation on a regular basis. Scats found on the routes leading to the feeding sites were also collected. Each scat was placed in a paper bag and the location and date of collection recorded. In this hot, arid environment, scats become dry a few hours after defecation and storage in paper bags keeps them dry and protected from condensation.
Genetic analysis
We extracted DNA from faecal samples using the protocol in Reed et al. (Reference Reed, Tollit, Thompson and Amos1997). Diagnostic mtDNA segments were used to confirm wolf DNA. We used 10 polymorphic wolf microsatellites to identify individuals. Details of the molecular procedures are outlined in Supplementary Information 1.
We used two population genetic approaches to examine whether a spatial correspondence exists between genetic and spatial grouping (i.e. INPA districts) of wolves. Details of the relatedness network analysis are provided in Supplementary Information 1.
Bayesian clustering
To assign individuals to populations we used STRUCTURE 2.3.1 (Pritchard et al., Reference Pritchard, Stephens and Donnelly2000), which uses multi-locus genotypes (X) to infer population structure. The Bayesian model assumes K populations that have different allele frequencies within a set of independent loci. We applied the admixture ancestry model once with the independent allele frequency model and once with the correlated model. The number of populations (K) was modified from 1 to 10. For each value of K 10 independent simulations were run for each of the two allele frequency models. Each simulation had a burn-in period of 500,000 iterations followed by a sampling of 500,000 iterations. The likelihood of K (i.e. Ln Pr(X|K)) and ΔK (Evanno et al., Reference Evanno, Regnaut and Goudet2005) were used to infer the more likely number of populations. This algorithm also calculates a membership coefficient (Q) for each individual in each population. We used the highest Q value to assign each individual to a population.
Population size estimation
We used three different approaches to estimate the wolf population size in the Negev: rarefaction curves, capture–mark–recapture, and a simple urn model called Capwire.
Rarefaction curve
In this technique the data are plotted as the number of unique multi-locus genotypes (i.e. individuals) as a function of sample size (number of scats analysed) and then fitted to a curve. The asymptote of the curve provides an estimate of the population size. Three fitting equations have been employed in similar previous studies: y = ax/(b+x) (Kohn et al., Reference Kohn, York, Kamradt, Haugt, Sauvajot and Wayne1999), y = a(1−e−bx) (Eggert et al., Reference Eggert, Eggert and Woodruff2003) and y = a−a(1−1/a)x, which was developed by D. Chessel (Frantz & Roper, Reference Frantz and Roper2006). In all equations a is the asymptote, b determines the rate of decrease of the slope, x is the number of scats sampled, and y is the number of unique genotypes. We used GIMLET to generate rarefaction curves (Valiere, Reference Valiere2002). Because the order in which DNA samples are added has an influence on the shape of the curve (Kohn et al., Reference Kohn, York, Kamradt, Haugt, Sauvajot and Wayne1999) we used R 2.10.1 (Ihaka & Gentleman, Reference Ihaka and Gentleman1996) to randomize (1,000 iterations) the order of the DNA samples and projected the asymptote for each of these randomizations using each of the three equations. The median of all 1,000 iterations of the asymptote was taken as an estimate of the population size (Frantz & Roper, Reference Frantz and Roper2006). The 95% confidence interval (CI) corresponds to the 0.025 and 0.975 percentiles.
Capture–mark–recapture (CMR) model
The study design (i.e. sampling at the feeding stations at monthly intervals) allowed us to easily adapt the genetic data to the CMR model. Identical multi-locus genotypes were considered to originate from the same individual and a history of captures and recaptures was assembled for each genotype. We divided the study period into 20 monthly sampling sessions. If the same genotype was sampled more than once in the same month we considered only one capture for that month. Analysis was performed with an open population model, using the POPAN module in MARK (White & Burnham, Reference White and Burnham1999). We used a set of four possible models: fully time dependent (pt,φt,bt) where p represents the probability of capture, φ represents the probability of an animal surviving between occasions and b is probability of entrance; constant catchability (p·,φt,bt); constant survival rate (pt,φ·,bt); and constant catchability and survival rate (p·,φ·,bt). We performed a goodness-of-fit using the RELEASE suite (Cooch & White 2007) to confirm that the most general sub-model adequately fitted the data. MARK uses AICc (Akaike's information criterion corrected for small sample size) for model selection.
Capture with replacement
In non-invasive sampling individuals may be ‘captured’ several times within one sampling session. When using classical CMR models such multiple captures must be pooled, as an animal should not be captured more than once in one session. This potentially wastes data. Capwire (Miller et al., Reference Miller, Joyce and Waits2005) uses a method developed to accommodate this type of data and is based on a simple urn model. There are two capture models: the even capture probability model (ECM) in which every individual is equally likely to be captured in each sampling session with a probability of 1/population size, and the two innate rates model (TIRM), which is more realistic, in which there are two types of individuals with different probabilities of capture. The software performs a likelihood-ratio test to determine which model is more suitable for the given data.
Results
Sample collection and microsatellite genotype
Over a period of 20 months from May 2007 to December 2008 a total of 334 faecal samples were collected and an additional three tissue samples were taken from wolves that had been shot by INPA rangers. Most scats (82%) were collected after the wolf control programme was terminated. Mitochondrial DNA for species identification was successfully amplified in 230 (69%) of the faecal samples, of which 95 were identified as wolf DNA, 93 were of red foxes Vulpes vulpes, 12 of dogs Canis lupus familiaris, nine of golden jackals Canis aureus and two of striped hyaenas Hyaena hyaena. In 5% of the samples that were identified, mtDNA of the prey (Ovis aries, Capra hircus, Bos taurus) rather than the predator was amplified.
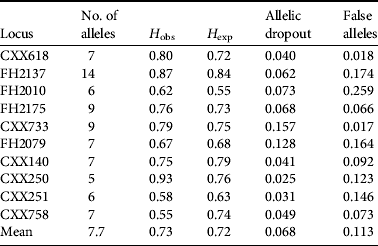
Of the 98 wolf DNA samples (i.e. 95 faecal samples plus three tissue samples) we obtained a complete or nearly complete multi-locus genotype for 76 samples. The mean rate of allelic dropout was 0.068 and mean rate of false alleles was 0.113 for all replicates at all microsatellite loci. The probability of two individuals possessing identical genotypes (PI) was 1.40 × 10−10 for a randomly mated population and 1.635 × 10−4 for siblings. Error rates and PI values of each locus were calculated using GIMLET (Table 1, Supplementary Information 1). We identified 52 unique haplotypes of which 12 were sampled more than once, with a mean of 1.46 samples per individual. The maximum resample rate was six (n=2).
Table 1 The number of alleles, observed (H obs) and expected (H exp) heterozygosity, and the rate of allelic dropout and false alleles for each of the 10 loci used in this study.
Population structure
We used the Bayesian model in the programme STRUCTURE. The highest Ln Pr(X|K) and ΔK were observed for K=3, indicating three genetic subpopulations (Figs 2 & 3a). This result was consistent for both correlated and independent allele frequency models. Most sampled individuals were easily assigned to a cluster as their estimated membership coefficient (Q) was > 0.7 for a specific population. However, three individuals (15, 7, and 11) could not be assigned because their Q values were ⩽ 0.7 (Fig. 3a).

Fig. 2 Number of subpopulations inferred by STRUCTURE. (a) Mean Ln Pr(X|K) over 10 runs for each K value. (b) ∆K for each K value, where ∆K=m|L(K+1)−2L(K)+L(K−1)|/s[L(K)]. Both Ln Pr(X|K) and ∆K were calculated for the correlated and independent models.

Fig. 3 Assignment of haplotypes by the STRUCTURE Bayesian algorithm. (a) Probability of belonging to each of three clusters (K=3) is denoted by shading (as in (b)). (b) Spatial distribution of haplotypes, assigned to the cluster with the highest probability. Empty circles are haplotypes that could not be assigned to a cluster. The frequency of each cluster (C1, C2, C3) in each of the districts is indicated by a bar chart. The heavy dashed line indicates the boundary between INPA districts.
The distribution of two of the genetic populations (C3 and C1; Fig. 3b) did not show any clear spatial separation between INPA districts. In Eilat district six individuals were of the C1 cluster and 10 of the C3 cluster, and in the South district 10 individuals were of the C1 cluster and eight of the C3 cluster (Fig. 3b). However, a third population (C2; Fig. 3b) had most individuals (n = 13) in the South district and only one individual in the Eilat district. Thus, the division between districts was significantly dependent on genetic clustering (χ22 = 8.114, P = 0.0172) because of the uneven distribution of the C2 cluster members between districts (Fig. 3b).
Population size estimation
The Kohn rarefaction equation gave the highest population estimate, of 176.8 wolves (95% CI 120.8–491.4), followed by that of Eggert with an estimate of 102.1 (95% CI 74.4–256.2), and Chessel with the lowest estimate of 86.2 wolves (95% CI 72.1–105.3; Table 2). The TIRM model of Capwire (L(TIRM)−L(ECM)=22.48) gave an estimate of 142 wolves (95% CI 89–179). For the CMR model the goodness-of-fit test confirmed that the most general sub-model adequately fitted the data (χ211 = 3.447, P = 0.983). The constant catchability and constant survival rate model ranked highest by AICc and gave an estimate of 173.4 wolves (95% CI 85.6–261.1; Table 3).
Table 2 Mean population size and mean density of the Arabian wolf Canis lupus arabs in the Negev Desert, and in the South and Eilat INPA districts separately (Fig. 1), based on five estimation methods (see text for details).

Table 3 Selection of MARK models (see text for details) for the Negev Desert population and INPA South and Eilat district populations separately.

1AIC value corrected for small sample sizes 2Difference in AICc between the focal and the best model 3AIC weight 4Model likelihood
A separate population estimation for each district showed that although the number of wolves in the Eilat district was considerably lower than in the South district, the density of wolves was similar in both districts based on the rarefaction estimates but higher in the South district based on the CMR and Capwire estimates (Table 2). CMR models showed that the lowest AICc value was obtained by the constant catchability model for the South district and the constant catchability and survival rate model for the Eilat district (Table 3).
Discussion
Our results show that the wolf populations in the two INPA districts had a similar genetic composition. The Bayesian model in STRUCTURE identified three genetic subpopulations: two distributed across both districts, and one mostly in the South district (Fig. 3). However about half of the individuals of the latter population were found near the south-eastern corner of the South district, at the border between the districts. Additional sampling would probably reveal their presence in the north-eastern corner of the Eilat district. Furthermore, network analysis of relatedness shows that individuals of both communities are present in both INPA districts in approximately equal proportions (Supplementary Fig. S1). Taken together, the evidence for considering the wolves in the two districts as separate, distinct populations is weak. Overall, our data suggest that all the wolves in the Negev Desert comprise a single population, which should therefore be managed as a single conservation unit. This view is supported by the extensive dispersal distance of Arabian wolves in the Negev (Hefner & Geffen, Reference Hefner and Geffen1999), implying that no isolation exists between wolf populations living throughout this region.
In these contradictory management approaches between the two INPA districts priority has been given to gazelles over wolves. This situation could have been avoided through a more balanced ecosystem approach that considers the ecological role of both species. The Arabian wolf is currently the top predator in this system, and the gazelle (mostly G. dorcas) is one of the largest native ungulates together with the Nubian ibex Capra ibex nubiana, onager Equus hemionus and Arabian oryx Oryx leucoryx. The latter two species were recently reintroduced into the Negev Desert. Both wolves and gazelles have key ecological roles in the Negev ecosystem (Mendelssohn & Yom-Tov, Reference Mendelssohn and Yom-Tov1999).
Does the Arabian wolf exert a significant impact on the gazelle population? This issue has never been systematically studied in the Negev Desert. Our findings indicate that the number of wolves in the southern Negev (i.e. Eilat district) is c. 30–50, whereas 60–100 inhabit the central Negev (i.e. South district). Both gazelle species have been counted annually in the Negev Desert since the 1970s. The mountain gazelle population is restricted to a single locality in Eilat district and has numbered c. ⩽ 40 since 1978 (Yom-Tov & Ilani, Reference Yom-Tov and Ilani1987; Dolev & Perevolotsky, Reference Dolev and Perevolotsky2004). Thus, the significance of mountain gazelle to the overall Negev ecosystem is negligible. However, the dorcas gazelle is widespread in the Negev, with densities reaching 5 km−2 in areas where many acacias are present and 0.1 km−2 in areas without acacias (Baharav, Reference Baharav1980). Based on total counts the population size of the dorcas gazelle in the Negev during 1980–1985 was estimated at 1,000–1,300 (Yom-Tov & Ilani, Reference Yom-Tov and Ilani1987; Mallon & Kingswood, Reference Mallon and Kingswood2001). During 1971–2009 the number of dorcas gazelles was 500–900 in the Eilat district (B. Shalmon, unpubl. data) and 150–550 in the South district (B. Shalmon & A. Tsoar, unpubl. data). Dorcas gazelle numbers in the Eilat district have fluctuated considerably in the past 40 years (Yom-Tov & Ilani, Reference Yom-Tov and Ilani1987); however, there are no comparable annual data on wolf numbers to support the claim that these fluctuations are related to predation. Nor is reliable data on predation of fawn or adult mountain and dorcas gazelle species available for the Negev population. Analysis of 777 wolf scats from the southern Negev (Shalmon, Reference Shalmon1986) revealed that the major dietary components of Arabian wolves are cow carrion (found in 62.5% of scats), fruit and vegetation (51.4% of scats) and human garbage (37.2% of scats). The frequency of indigenous prey, including gazelles, in the scats was only 6.3%. In North America Messier et al. (Reference Messier, Carbyn, Fritts, Seip, Carbyn, Fritts and Seip1995) showed that the number of wolves per area is linearly related to the density of deer-sized prey. In these populations, wolves consumed mainly large ungulates (northern Minnesota, Kunkel & Mech, Reference Kunkel and Mech1994; Isle Royale National Park, Vucetich et al., Reference Vucetich, Peterson and Schaefer2002; Yellowstone National Park, Wright et al., Reference Wright, Peterson, Smith and Lemke2006) In contrast, the Arabian wolves in the Negev seem to be dependent on human-related waste and domestic livestock and seldom rely on natural prey.
The implications of our results are that a major change in wolf management in both INPA districts is required. Firstly, Arabian wolves should not be culled without a robust corresponding study on the effects of such action. The Arabian wolf is a threatened species, living in an arid zone with limited resources (Dolev & Perevolotsky, Reference Dolev and Perevolotsky2004; Sillero-Zubiri et al., Reference Sillero-Zubiri, Hoffmann and Macdonald2004). Its population fluctuates similarly to other species dwelling in this region, and may reach critical low levels. Furthermore, culling is indiscriminate and could thus inadvertently eliminate a distinct genotype and thereby reduce the evolutionary potential for survival of this population. Secondly, we suggest that reducing access to human-related waste and domestic livestock could adjust the wolf population size to this arid environment's natural carrying capacity by forcing the wolf population to feed more on natural prey. Thirdly, the effect of droughts and habitat modification by human activity on the dorcas gazelle population in the Negev should be systematically investigated before further culling of predators is carried out. At present, both districts are modifying their management policies with respect to wolves. Wolf culling has been terminated, and new programmes are being developed for limiting the access of carnivores to human waste and agriculture surplus. We have also developed a study on wolf resource utilization, using GPS collars, to understand the dependency of wolves on human resources. Finally, we call for more detailed study on the population dynamics of gazelles in the Negev, especially in relation to effects of global warming, before instigating any further predator control programmes in this region.
Acknowledgements
We would like to thank Shirley Bar-David, Naomi Paz, Uri Roll, Benny Shalmon and Yoram Yom-Tov for constructive comments, and Nimrod Ben-Aharon, Mark Catz, Ben Drori, Gil Evron, Ariela Gotlieb, Reuven Hefner, Yoram Hemo, Tzoor Magen and Enav Vidan for maintaining the feeding sites and assisting in the collection of scats. The Israel Nature and Parks Authority provided logistic and financial support.
Biographical sketches
Orly Cohen is interested in applying molecular approaches to ecological and conservation-related problems. Adi Barocas is a behavioural ecologist interested in the effects of social integration and complexity on survival. He is also interested in conservation and management of carnivores. Eli Geffen has broad, global interests in behaviour, genetics and conservation of canids.








