Introduction
The lichenized genus Allocetraria Kurok. & M. J. Lai was described in 1991, with a new species A. isidiigera Kurok. & M. J. Lai, and two new combinations: A. ambigua (C. Bab.) Kurok. & M. J. Lai and A. stracheyi (C. Bab.) Kurok. & M. J. Lai (Kurokawa & Lai Reference Kurokawa and Lai1991). The main distribution area of Allocetraria species was reported to be in the Himalayas, including China, India, and Nepal.
Allocetraria is characterized by dichotomously or subdichotomously branched lobes and a foliose to suberect or erect thallus with sparse rhizines, angular to sublinear pseudocyphellae, palisade plectenchymatous upper cortex, as well as producing usnic acid but never atranorin (Kurokawa & Lai Reference Kurokawa and Lai1991). It is a well-supported monophyletic group within the cetrarioid clade in Parmeliaceae (Saag et al. Reference Saag, Randlane, Thell and Obermayer2002; Thell et al. Reference Thell, Högnabba, Elix, Feuerer, Kärnefelt, Myllys, Randlane, Saag, Stenroos, Ahti and Seaward2009; Nelsen et al. Reference Nelsen, Chavez, Sackett-Hermann, Thell, Randlane, Divakar, Rico and Lumbsch2011). Ten species of Allocetraria have been accepted in the genus worldwide; China is the main distribution area of the genus, as all ten species have been reported there (Kurokawa & Lai Reference Kurokawa and Lai1991; Thell et al. Reference Thell, Randlane, Kärnefelt, Gao, Saag, Daniels, Schulz and Peine1995; Randlane et al. Reference Randlane, Saag and Obermayer2001; Wang et al. Reference Wang, Wang and Wei2014). During our taxonomic study of Allocetraria, a new species was found.
Materials and Methods
A dissecting microscope (ZEISS Stemi SV11) and compound microscope (ZEISS Axioskop 2 plus) were used to study the morphology and anatomy of the specimens. Colour test reagents [10% aqueous KOH, saturated aqueous Ca(OCl)2, and concentrated alcoholic p-phenylenediamine] and thin-layer chromatography (TLC, solvent system C) were used for the detection of lichen substances (Culberson & Kristinsson Reference Culberson and Kristinsson1970; Culberson Reference Culberson1972).
Nineteen fresh specimens were chosen for DNA extraction (Table 1), in which eight species of Allocetraria were included. The remaining three species of the genus, A. capitata R. F. Wang et al., A. denticulata (Hue) A. Thell & Randlane and A. isidiigera, are absent because of a lack of fresh specimens and corresponding sequences in NCBI. The extraction procedure followed the modified CTAB method (Rogers & Bendich Reference Rogers, Bendich, Gelvin and Schilperoort1988). PCR amplifications were performed using a Biometra T-Gradient thermal cycler. The primer pairs ITS1 (White et al. Reference White, Bruns, Lee, Taylor, Innis, Gelfand, Sninsky and White1990) and LR1 (Vilgalys & Hester Reference Vilgalys and Hester1990) were used to amplify the nrDNA ITS regions. Twenty-seven sequences were aligned with the program MEGA5 (Tamura et al. Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011); 19 specimens were sequenced by the authors, and 8 sequences were downloaded from GenBank, including outgroups Tuckermanopsis ciliaris (Ach.) Gyeln., Usnocetraria oakesiana (Tuck.) M. J. Lai & J. C. Wei, and Vulpicida juniperinus (L.) J. E. Mattsson & M. J. Lai. The phylogenetic analysis was executed with MEGA5. Model TN93+G was set according to the lowest BIC scores (Bayesian Information Criterion). The Maximum Likelihood (ML) method was used in constructing the phylogenetic tree and the reliability of the inferred tree was tested by 1000 bootstrap replications.
Results and Discussion
The ML tree (Fig. 1) based on the ITS sequences demonstrates that the eight species of Allocetraria clustered into a moderately well-supported (86% bootstrap value) monophyletic clade. Allocetraria yunnanensis R. F. Wang et al. is well clustered (94% bootstrap value) together with the similar species A. sinensis X. Q. Gao as a separate group, despite their obvious differences. Therefore A. yunnanensis is a new species, well supported by DNA data.
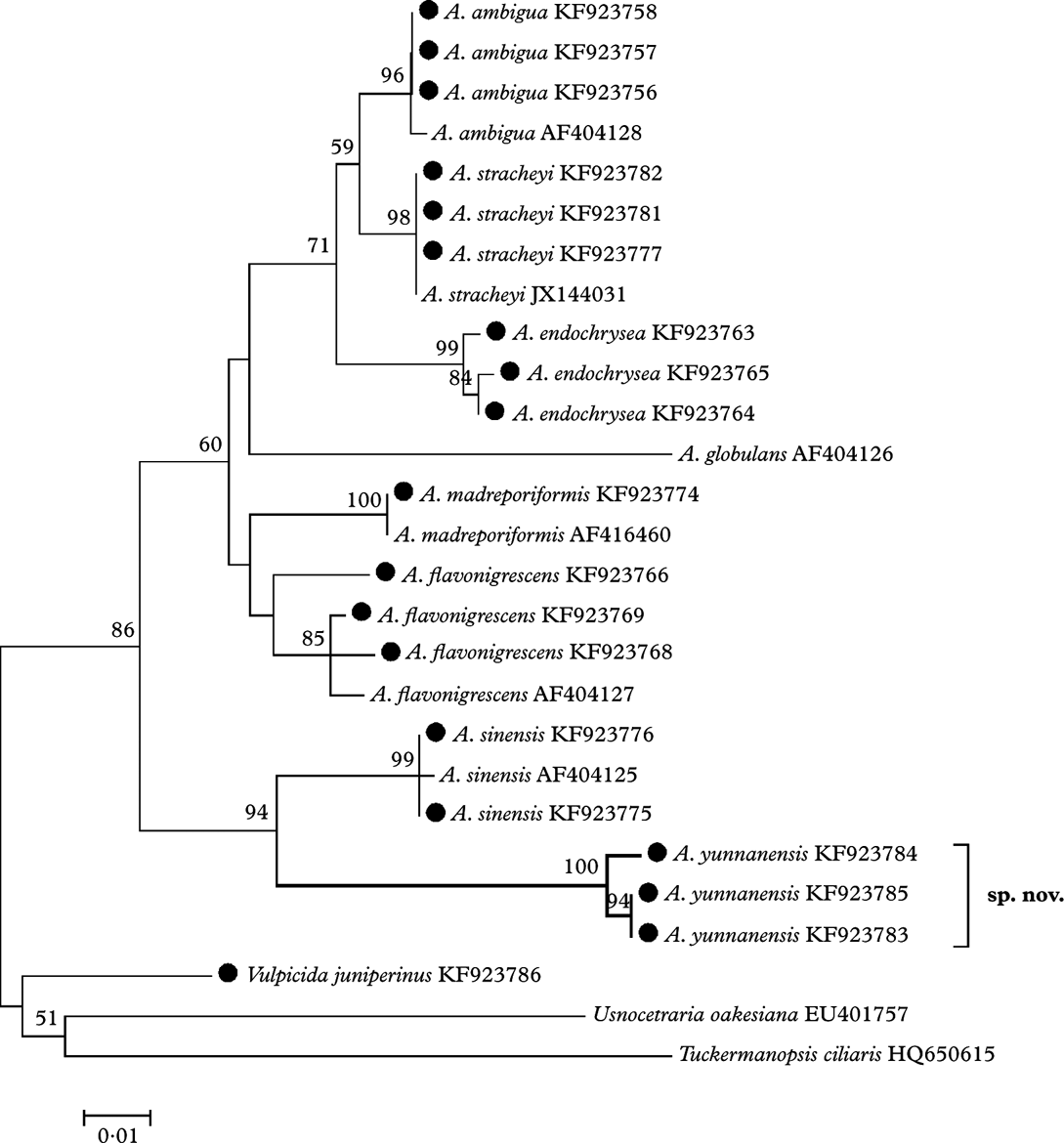
Fig. 1. The ML tree based on nrDNA ITS region sequences, the specimens marked with ‘•’ were examined by the authors. Nucleotide: TN93+G model, bootstrap=1000. Genetic distance scale=0·01. The number at each node represents bootstrap support value (numbers lower than 50 are not shown).
The New Species
Allocetraria yunnanensis R. F. Wang, X. L. Wei & J. C. Wei sp. nov.
MycoBank No.: MB809069
Similar to A. sinensis in habitus, but differs from the latter and all other members in the genus by having a shiny upper surface and a lower surface which is strongly wrinkled and has marginal pseudocyphellae present on the lower side in the form of a white continuous line or spot.
Type: China, Yunnan Province, Deqin County, Meli Village, Meili Snow Mountain, on soil, 28°38·191′N, 98°36·304′E, alt. 4800 m, 10 September 2012, R. F. Wang YK12012 (HMAS-L128218—holotype).
(Fig. 2)

Fig. 2. Allocetraria yunnanensis R. F. Wang, X. L. Wei & J. C. Wei, sp. nov. (holotype). A, habit; B, anatomy of apothecium in vertical section; C, longitudinal section of the thallus; D, pycnoconidia. Scales: A=1 cm; B–D=20 μm.
Thallus foliose, suberect to prostrate, dorsiventral, caespitose, 1·0–1·5 cm high. Lobes narrow, 1–4 mm wide, 200–350 µm thick, sublinear-elongate, flat to slightly concave, irregularly branched lobules. Upper surface greenish yellow to yellow or green, smooth, slightly shiny. Lower surface white to brown or almost dark brown, strongly wrinkled, dull. Pseudocyphellae located on the ridges of the lower surface, forming a continuous line or spot. Rhizines marginal on the lower side, sparse, simple, dark, 1–2 mm long. Epicortex 10–15 µm thick, non-cellular; upper cortex 30–60 µm thick, more or less palisade plectenchymatous; lower cortex 35–50 µm thick, more or less palisade plectenchymatous. Medulla light yellow to yellow.
Apothecia rare, terminal or marginal, up to 6 mm diam., with brown disc; thalline margin rather thick and crenulate. Asci narrowly cylindrical 35–50×10–15 µm; ascospores globose, 5–9 µm diam., or subglobose, 6–8×5–7 µm.
Pycnidia marginal, frequent, black, on emergent projections; pycnoconidia filiform, one end slightly swollen, 10–18×1·0–2·5 µm.
Chemistry
Lichesterinic, protolichesterinic, secalonic and usnic acids; cortex K−, KC+ yellow; medulla K−, C−, KC−, PD−.
Etymology
The epithet ‘yunnanensis’ is derived from the locality of the new species ‘Yunnan’, a province of China. Known only from the type locality.
Ecology and substratum
On the ground, mixed with moss.
Specimens examined. China: Yunnan Province: Deqin County, Meli Village, Meili Snow Mountain, on soil, 28°38·191′N, 98°36·304′E, alt. 4800 m, 2012, R. F. Wang YK12004 (HMAS-L-128219), YK12018 (HMAS-L-128220).
This project was supported by the National Natural Science Foundation of China (31200018) and the Main Direction Program of Knowledge Innovation of the Chinese Academy of Sciences (KSCX2-EW-Z-9). We thank Ms. H. Deng for providing considerable assistance during the studies in HMAS-L.




