Introduction
Although the first citizen science activity is traditionally considered to be the Christmas Bird Count of the Audubon Society in 1900 (Butcher et al. Reference Butcher, Fuller, McAllister and Geissler1990), public participation in acquisition of knowledge aimed at scientific research is a process that has only relatively recently increased in popularity. Coined as a description of a laboratory activity focused on the analysis of data on birds, collected over the years by a large number of amateurs (Bonney Reference Bonney1996), the expression ‘citizen science’ is currently used as an umbrella term for all approaches that entail the involvement of citizens and researchers in collaborative research activities.
In the last 25 years, thousands of projects have been organized involving millions of participants in the collection and/or processing of data all over the world, covering a wide range of domains. According to a survey focused on European citizen science projects (Hecker et al. Reference Hecker, Garbe, Bonn, Hecker, Haklay, Bowser, Makuch, Voge and Bonn2018), 75.7% of the projects were related to life sciences, 11% to humanities and social sciences, 7.5% to natural sciences, 5.8% to engineering disciplines.
Citizen science is becoming an increasingly well-developed and valued approach, even at an international political level (e.g. Bio Innovation Service 2018; Turbé et al. Reference Turbé, Barba, Pelacho, Mugdal, Robinson, Serrano-Sanz, Sanz, Tsinaraki, Rubio and Schade2019). For example, when considering the research and innovation funding programme Horizon Europe, citizen science is recognized as a strategic practice to strengthen European citizens’ trust in research and in its achievements and to raise the level of scientific literacy.
Among the most successful tools for collecting data in citizen science initiatives for natural science, iNaturalist (www.inaturalist.org/) is one of the most popular worldwide (Aristeidou et al. Reference Aristeidou, Herodotou, Ballard, Young, Miller, Higgins and Johnson2021). It is an online platform built, as a joint initiative of the California Academy of Sciences and the National Geographic Society, on the concept of mapping and sharing observations of biodiversity across the globe, as well as for seeking identification help from the community of users. iNaturalist uses artificial intelligence (computer vision systems) trained on photos and identifications uploaded on the platform itself to provide automated taxon identification suggestions. That potentially allows everyone to associate a putative name to an observed organism, without specific knowledge of the taxon.
iNaturalist has become very common and widely used: today, the iNaturalist community has more than two million registered users, who have made over 120 million observations of 400 000 species. Following registration on this social platform, available for PC and mobile devices, individuals can upload observations and identify organisms. Observations are reports of the occurrence of one organism in a given space and time, which can be supported by some media (currently images or sound) tagged with metadata such as taxonomic identification, date, associated text, tags, and geographic information. The community (both experts and non-experts) can additionally add identifications of the uploaded observation. When two or more observers agree on the identification, its status moves from ‘Needs ID’ to ‘Research-grade’ (RG), where RG is the highest quality level in iNaturalist for an identification at a level lower than family. Moreover, RG observations are also aggregated in the Global Biodiversity Information Facility database (GBIF; www.gbif.org) by default.
These observations, which provide large-scale, real-time data on biodiversity, can also represent valuable resources for scientific research. Citizen science platforms are becoming a popular tool for researchers allowing scientists to access a wider data collection network (Follett & Strezov Reference Follett and Strezov2015). For easily identifiable groups, iNaturalist can be a valuable source of information on distribution, enabling it to be used to supplement traditional survey methods (Jackson et al. Reference Jackson, Gergel and Martin2015). iNaturalist data have been used in a number of scientific studies for a wide range of purposes including tracking changes in species distribution both of common and elusive species (Ricca & Cheung Reference Ricca and Cheung2021; Rosa et al. Reference Rosa, Cavallari and Salvador2022), monitoring the spread of alien-invasive species (Creley & Muchlinski Reference Creley and Muchlinski2017; Hiller & Haelewaters Reference Hiller and Haelewaters2019), studying the impacts of urbanization and land-use change on biodiversity (Lee et al. Reference Lee, Kahal, Kinas, Randall, Baker, Carney, Kendell, Sanderson and Duke2021), and analyzing plant phenology (Barve et al. Reference Barve, Brenskelle, Li, Stucky, Barve, Hantak, McLean, Paluh, Oswald and Belitz2020) and its links with climate change (Iwanycki Ahlstrand et al. Reference Iwanycki, Primack and Tøttrup2022). The use of iNaturalist data is also increasing in studies on rare or endangered species both for monitoring and tracking the effects of conservation efforts (Wilson et al. Reference Wilson, Pan, General and Koch2020) and even to study the effects of recent pandemic events (Vardi et al. Reference Vardi, Berger-Tal and Roll2021). Citizen science and iNaturalist observations have been used to detect species previously unknown to science (Winterton Reference Winterton2020) and to explore species occupancy and distribution in areas with no systematic research institutions and scarce funding for science (Wangyal et al. Reference Wangyal, Bower, Vernes and Thinley2022).
While the idea of mapping biodiversity and sharing observations is to be commended and the storage of open access data on a free platform is a powerful tool both for citizen science and research, we must consider that species identification is not an easy task. Developing the skills to identify species of any taxon can require years of practice and, often, instruments and products that are not within everyone's reach (i.e. microscopes, chemicals, chromatography equipment, literature).
In 2022, about one million observations of lichens were available worldwide on iNaturalist, which is less than 1% of the total uploaded observations, meaning lichens are still under-represented on the platform.
In lichenology, most of the species require microscopic observations and chemical tests and, even for experts, images are barely sufficient to reach a correct identification at the species level. Therefore, having thousands of lichen records reported by non-lichenologists and identified via computer vision (a type of artificial intelligence) raises concerns among the scientific community (McMullin & Allen Reference McMullin and Allen2022). In this work, we evaluated the effectiveness and reliability of lichen identification on iNaturalist comparing the results of lichen identification performed by iNaturalist users using the platform with results obtained by expert lichenologists.
Material and Methods
Test concept
The comparison between iNaturalist users and expert lichenologists was organized in well-defined areas of comparable size (FCUL campus in Lisbon, Grugliasco University campus in Turin, and Botanical Garden and Villa Giulia in Palermo) in order to identify the different sources of variability and error in lichen recognition. The expert assessments aimed to provide the baselines against which to compare the iNaturalist users’ performances and were not intended as replicates of the same experimental design.
The lichenological information on the sites, already on the platform, indicated significant variation between them. Within a radius of 50 km from the areas, the following data were available: 212 taxa by 933 observations from the area surrounding Lisbon; 82 taxa by 438 observations from the area surrounding Turin; 11 taxa by 20 observations from the area surrounding Palermo.
iNaturalist users received only standard information on using the platform through seminars included in specific projects at each site (+Biodiversidade@CIÊNCIAS in Lisbon, DIVERSAGROVET project in Turin, and activities organized under the guidance of the Italian Botanical Society, regional sections ‘Sicilia - SBISI’ and ‘Piemonte e Valle d'Aosta’, and Lichenology Working Group in Palermo). No specific information about lichen taxonomy was provided to the iNaturalist users.
‘Tank Projects’ for iNaturalist users
So-called ‘Tank Projects’ are three main citizen science projects carried out in Portugal and Italy. +Biodiversity-@CIÊNCIAS is a project funded by the Faculty of Sciences of the University of Lisbon (FCUL) with the aim of contributing to sustainability within the FCUL campus and in its interaction with the city of Lisbon (Portugal). The project began during 2020–21, aiming at increasing citizens’ awareness of the importance of biodiversity. To involve the scientific community and the citizens who live and work in the area, the online project ‘BioDiversity4All’ (https://www.biodiversity4all.org) was created on the iNaturalist platform for data recording and monitoring.
The area selected for the activity extends over 160 ha, of which c. one quarter has tree cover and includes the FCUL campus, a public garden with a small artificial lake, the hippodrome and the sports complex of the faculty with an associated green area. The initiative was extremely successful, and the online platform so far has gathered over one million observations, referring to more than 10 000 species, with at least 8000 registered users and more than 11 000 followers on Facebook.
The DIVERSAGROVET project was conceived in 2021 by the Department of Agricultural, Forest and Food Sciences of the University of Turin (Italy) with the goal of raising citizen awareness of the value of biodiversity, allowing free access to the green areas surrounding university facilities. To map and monitor the biodiversity of the Grugliasco University campus, a specific data collection project was launched on the platform iNaturalist (https://www.inaturalist.org/projects/diversagrovet) involving students, citizens, and academic staff who work on the campus or live nearby.
Monitoring activities were performed on a 24-ha area, that includes green areas with both wild and cultivated plants, greenhouses, agricultural areas (orchards, vineyards and fields), a livestock farm and a small hill.
The SBISI activity was included among those proposed in 2022 by the regional sections ‘Sicilia - SBISI’ and ‘Piemonte e Valle d'Aosta’ and by the Lichenology Working Group of the Italian Botanical Society. The main purpose of this activity was to become familiar with the iNaturalist platform for collecting lichenological data. The selected areas were two contiguous urban parks: the Botanical Garden and Villa Giulia. The Botanical Garden covers an area of more than 10 ha and hosts 1692 plant species within the collections (http://ortobotanico.unipa.it/collezioni.html). Mediterranean, tropical and subtropical plants are represented, in large collections, as well as by numerous specimens of exotic plants. Villa Giulia, a typical Italian garden, is a public park of 7 ha. In both parks there are several stone cultural heritage artworks.
Data collection
Data collection by SM, DI and SR was carried out by expert lichenologists with many years of experience in two steps: identification of the species in the field and checking dubious specimens in the laboratory.
The field activity lasted a total of 8–10 hours for each study area. On 5–7 July 2021, SR and SM carried out a survey of three half days in the FCUL campus in Lisbon, while in Turin, DI carried out a two-day survey (19 July and 30 September 2022) on the Grugliasco campus, and in Palermo SR dedicated half a day to each of the two parks (11 May 2022 at the Botanical Garden and 27 October 2022 at Villa Giulia).
Although the surveys were not intended to document all of the lichen species in the study areas, they were able to identify the most prevalent ones in the locations where project participants detected lichen occurrence. Identification and nomenclature were mainly based on the online keys published in ITALIC (Nimis & Martellos Reference Nimis and Martellos2022), Clauzade & Roux (Reference Clauzade and Roux1985) and Smith et al. (Reference Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009).
Data mining and analysis
Data belonging to the projects ‘BioDiversity4All’, ‘DIVERSAGROVET’ and SBISI were downloaded from the iNaturalist platform selecting the category ‘Fungi including lichens’ for the period prior to the surveys made by the three expert lichenologists. All the records collected before the experts surveyed the areas were checked to select those referring to lichens, looking at the images when the taxonomic rank reported in the platform was insufficient to retrieve the information. When corrections were made by other users, the original identification of the first observer was considered.
Finally, a comparison was made between the species lists produced by the experts and the iNaturalist users. Taxa which were reported by experts and were not reported by iNaturalist users were investigated to determine potential issues that prevented their identification by the iNaturalist users. For species only reported by iNaturalist users the images were analyzed to evaluate, when possible, the accuracy of their identification. When images were not sufficiently clear, the ecology and distribution of the suggested species were considered to assess the reliability of their occurrence.
Results
In the three study areas, 108 species were inventoried (Table 1). The number of species that required verification in the laboratory was 13 in Lisbon, 8 in Turin and 5 in Palermo.
Table 1. A list of lichen species recorded by experienced lichenologists (Exp), iNaturalist users (iNat) and both (Both) in Lisbon, Turin and Palermo. * = species identified after laboratory verification by experienced lichenologists.
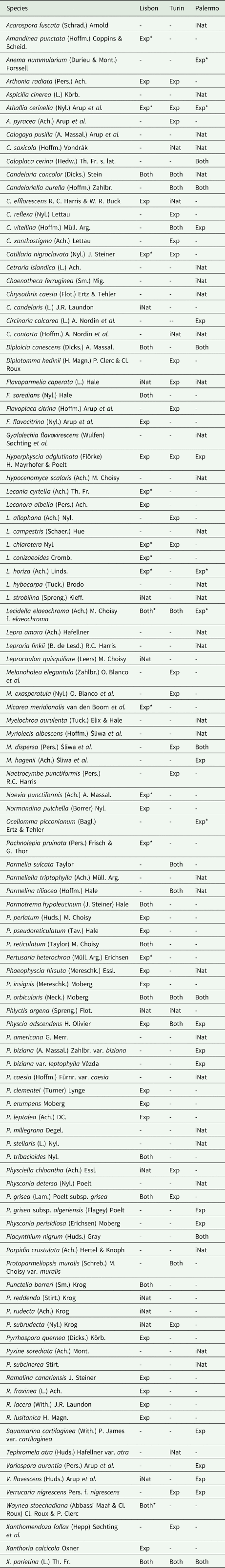
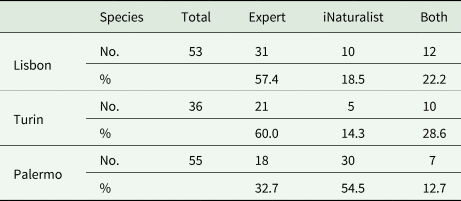
The experienced lichenologists (herein known as ‘experts’) identified almost 60% of the species in Turin and Lisbon, but only 33% in Palermo, when the number of species is considered but not the accuracy of the identification (Table 2). The overlap between the experts and iNaturalist users in Palermo was similarly lower, indicating a higher level of error in the users’ identifications. This can be attributed to a more limited iNaturalist lichen dataset available for that area (only 20 observations were available prior to this study for Palermo and surroundings).
Table 2. Summary of the total number and percentage of species found by experts (i.e. experienced lichenologists), iNaturalist users (species identity not checked or confirmed by experts) and both, in the three collection sites.
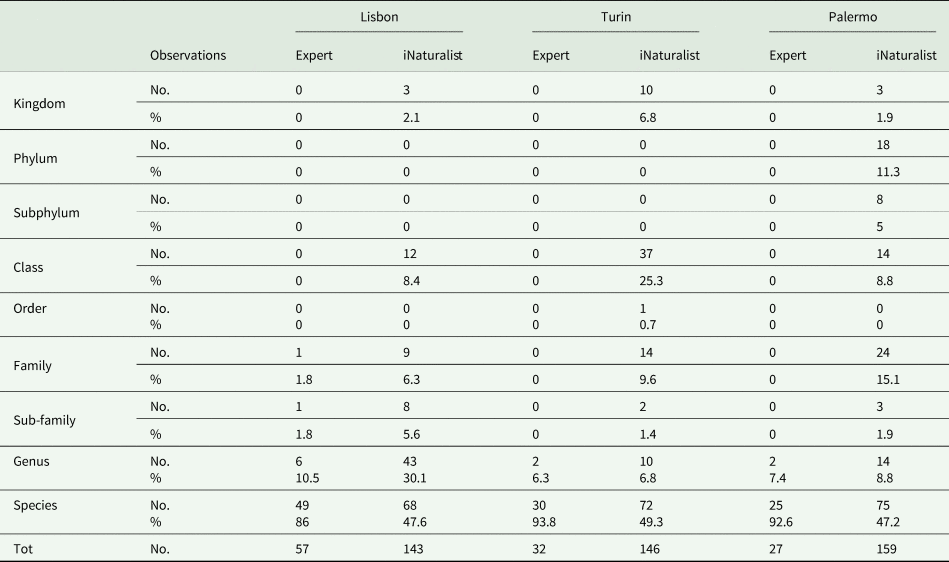
In all cases, more than 85% of the observations by experts led to identification at the species level and around 10% at the genus level. Conversely, less than 50% of the specimens were identified at the species level by iNaturalist users, more than 50% of identifications being referred to higher taxonomic ranks (Table 3).
Table 3. Summary of the total number and percentage of observations grouped by taxonomic rank found by experts (i.e. experienced lichenologists) and iNaturalist users in the three collection site.
Macrolichens (lichens with physical features easily seen with the naked eye) were over-represented in iNaturalist. Among the taxa which were reported only by experts, about 52%, 64% and 60% were microlichens (i.e. crustose lichens with morphological features that require microscopic observations for identification) in Lisbon, Turin, and Palermo, respectively. A sizeable percentage of overlooked species belonged to Physciaceae (Lisbon 23%, Turin 14% and Palermo 26%), while the rest included several genera and species (Fig. 1, Table 1).
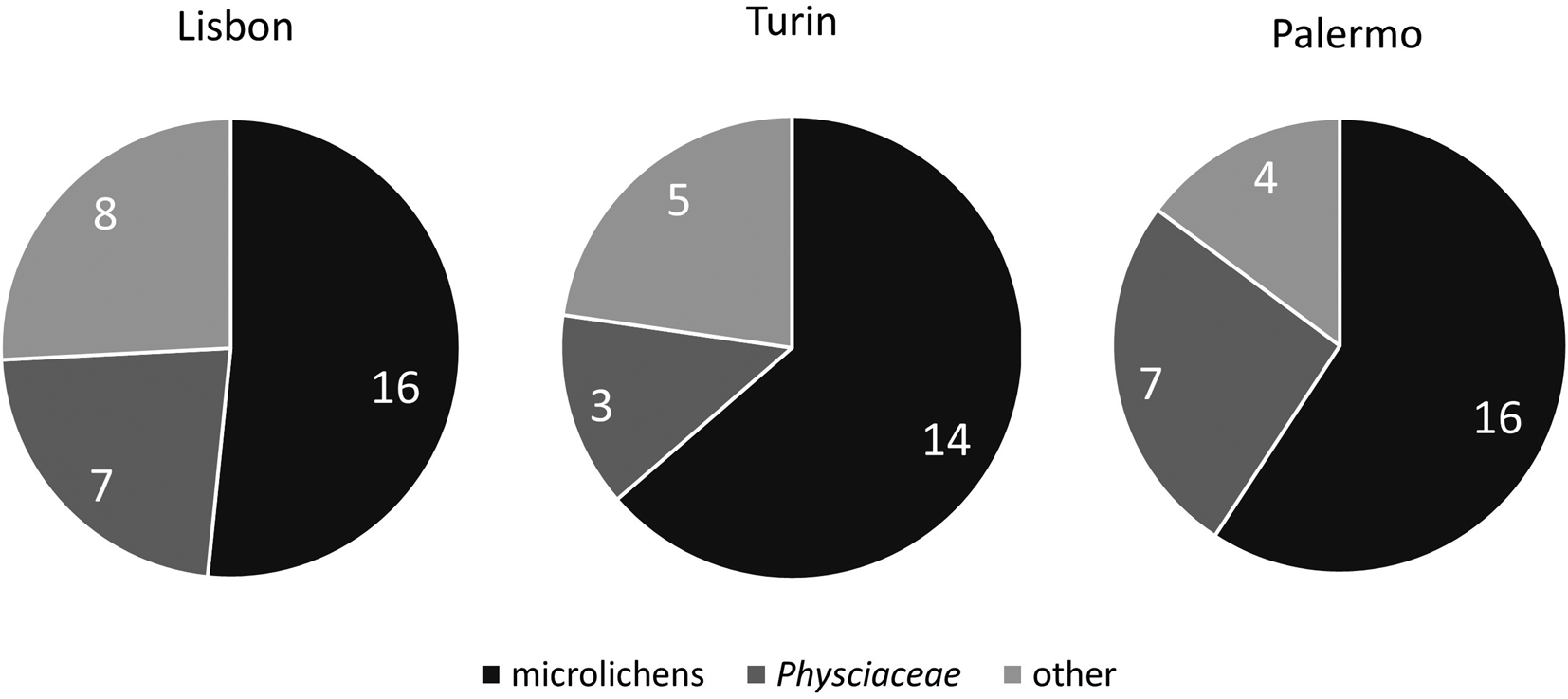
Figure 1. Proportion of overlooked lichen species belonging to Physciaceae in relation to groups at the three sites.
Of the identifications at the species level exclusively reported on iNaturalist, 70% were incorrect. When images were available, while it was not always possible to accurately identify the species, it was often possible to exclude the suggested identification based on diagnostic characters. Additionally, based on the ecology and distribution of the proposed species, 7% of the identifications were considered likely to be correct, while another 7% were considered unreliable.
Discussion
iNaturalist is an easy-to-use platform that can offer an effective learning tool to citizens (Chozas et al. Reference Chozas, Nunes, Serrano, Ascensão, Tapia, Mágues and Branquinho2023). It also comes at no cost, making it accessible to everyone. The platform's appeal is that it is accessed via mobile devices, and in particular smartphones, making it particularly appealing to the younger generation.
Our findings support the conclusions reached by McMullin & Allen (Reference McMullin and Allen2022), cautioning against the use of unchecked data obtained from the platform in lichenology, demonstrating substantial inconsistency between results gathered by iNaturalist users and experts.
Experts dealing with tiny and taxonomically difficult organisms like fungi and insects know that species identification based on images is either impossible or at least highly uncertain (Casanovas et al. Reference Casanovas, Lynch and Fagan2014; Prudic et al. Reference Prudic, Oliver, Brown and Long2018). Our results showed that this is as true for artificial intelligence as it is for humans. In particular, three distinct sources of uncertainty can be identified in the use of artificial intelligence for lichen identification. Firstly, the need to examine microscopic and chemical characteristics prevents the identification of entire lichen genera in situ. During the three surveys, experts had to complete their analysis in the laboratory, supplementing the species list and identifications established during field work. This constraint was also noted by de Groot et al. (Reference de Groot, Pocock, Bonte, Fernandez-Conradi and Valdés-Correcher2023) in their analysis of citizen science initiatives aimed at surveying arthropods in forested areas.
Several lichen species have such small and inconspicuous thalli, consisting of millimeter-sized granules/squamules or having a similar colour to the substrate, that they are almost imperceptible to the naked eye. The only indication of their occurrence may be through the presence of minute apothecia which can only be detected by experienced or highly perceptive individuals. This results in a significant underestimation of microlichens. A detection bias affecting iNaturalist observations with lichens was noted also for birds (Callaghan et al. Reference Callaghan, Poore, Hofmann, Roberts and Pereira2021). Furthermore, in urban areas, where citizen science activities are more frequent but where pollution poses a significant challenge to the growth of sensitive species, poorly developed thalli may lack the morphological features necessary for accurate identification. Species that are very similar in colour, shape and size, but differ in small details or specific structures mostly ignored by iNaturalist users tend to be underrepresented, as demonstrated in this study with the Physciaceae family. A similar situation was reported by Koo et al. (Reference Koo, Oh, Park and Im2022) when non-experts were not able to discern two similar species of amphibians based on the external shape alone. Therefore, inherent characteristics of lichens present challenges in the application of iNaturalist to lichenology.
Secondly, the current functioning of the platform does not guarantee a high level of accuracy in identification. The ‘research grade’ designation is given when at least two users independently agree on the identification, regardless of their level of expertise, and there are no means of revision against it. Consequently, the accumulation of two errors in an event that can occur with high probability will result in an RG misidentification in iNaturalist records, if not properly scrutinized.
Additionally, the low quality of some images and the absence of microscopic and chemical characters make it challenging, or even impossible, for experts to provide accurate evaluations. The lack of physical specimens stored in herbaria further hinders assessment of the accuracy of the identification (McMullin & Allen Reference McMullin and Allen2022). Although the practice of adding images concatenated to that of the organism portrayed as a species (e.g. details, microscopic characters or images depicting their habitat and ecology) is extremely helpful in the scrutiny (Table 4), it is still an uncommon practice.
Table 4. Assessment of the accuracy (likely, unlikely and wrong) of species identification by iNaturalist users as determined by the analysis of the associated imagery, ecology and distribution of the identified species.
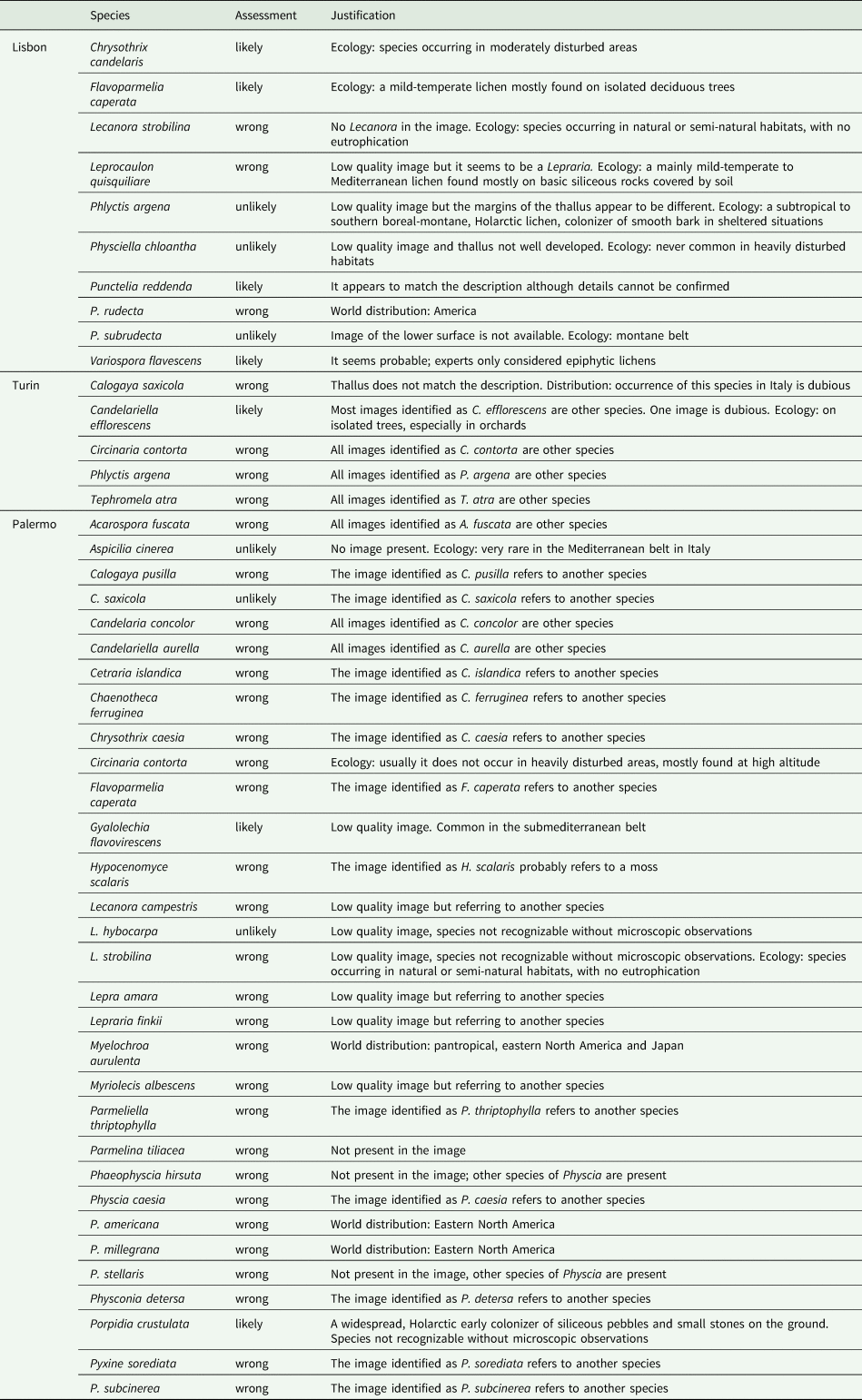
Without a strong dataset of precise identifications in a given area, as in the case of Palermo, the computer-generated suggestions may refer to species that are morphologically similar but not occurring in the area because the algorithm is not designed to take ecological factors into account when providing tentative identifications. Finally, dishonesty cannot be prevented in a system that is open access and free by definition. Those individuals who are determined to increase their personal record of observations, might blindly guess or put too much trust in the machine learning. Machine identification works well with well-known species with large training sets, but it is unreliable for scarcely represented groups like lichens, or where background data are lacking (https://www.inaturalist.org/pages/computer_vision_demo). Even if strongly discouraged in the iNaturalist guidelines, inexperienced users tend to agree too quickly with given identifications, despite knowing almost nothing about the species.
With specific regard to the platform, some measures could help improve the possibility of posting and handling data for research purposes. A crucial point is that users should be aware of the key characters for identifying lichens. Even just taking macrolichens into consideration, there are several useful tools that could be developed, such as introductory guides, tutorials on what have to be included and documented in a useful image (e.g. important morphological features, vegetative and reproductive characters, etc.), and best practices for gathering valuable data from iNaturalist photographs. Furthermore, for the purpose of a more accurate observation, the use of a ×10 magnifying lens could be encouraged. McMullin & Allen (Reference McMullin and Allen2022) proposed a set of recommendations for documenting observations of lichens, which can also be adapted to other challenging taxa, and iNaturalist users have already taken the initiative to gather and compile online resources for this purpose. On the other hand, researchers should be facilitated in the review process of iNaturalist data. For example, creating a specific filter to separate lichens from other fungi would save a lot of time. Currently, two different phyla (Ascomycota and Basidiomycota) and various classes that do not include only lichens must be searched to obtain a dataset containing lichens. This requires labour intensive sorting and since specific filtering does not currently exist, verification must be done by checking manually all the taxa. Furthermore, considering the robustness of the data for statistical processing, redundant data should be highlighted (i.e. images of the same specimen taken by different users). In this view, also a higher agreement threshold to determine what is/is not ‘researchable’ should be reached and provided. Similarly, a higher agreement threshold for determining what is/is not ‘research grade’ could provide more reliable data. For instance, it would be useful to add ‘redundant’ identifications to records that are already RG (in fact data users can choose to limit the data set to observations with three or more confirmations, this could lead to a better population to work through).
Conclusions
iNaturalist is potentially an exceptional tool for citizen science and outreach efforts in lichenology, as shown by the increase in lichen records on the platform. With their year-round occurrence on trees, soil and rocks, and their diverse shapes and colours, lichens are very attractive for projects on biodiversity to counteract ‘plant blindness’.
However, due to inherent uncertainties in lichen identification, by untrained or poorly trained volunteers who also mostly cannot access professional tools for correct identification, its application for research purposes is currently limited. While well-defined tasks involving a few identifiable species (e.g. in our study areas Xanthoria parietina, Diploicia canescens) could be successfully carried out using iNaturalist data, relying on this platform for extensive biodiversity surveys is not recommended. Actually, these activities would increase the risk that some data achieve RG identification and are included in GBIF erroneously. While data collection for large areas with small sampling effort may be useful, it is crucial to generate high-quality data and train users to confidently incorporate observations into research activities.
Acknowledgements
This work received support from the EU through the Erasmus+ Programme and from the grant UIBD/00329/2020 for the Centre for Ecology, Evolution and Environmental Changes (cE3c) from FCT. SR acknowledges the support of NBFC to the University of Palermo, funded by the Italian Ministry of University and Research, PNRR, Missione 4 Componente 2, ‘Dalla ricerca all'impresa’, Investimento 1.4, Project CN00000033. The authors warmly thank Lucy Sheppard for language revision.
Author ORCIDs
Silvana Munzi 0000-0002-0071-7699; Deborah Isocrono, 0000-0003-0570-0674; Sonia Ravera 0000-0001-5223-7964.
Competing Interests
The authors declare none.








