Published online by Cambridge University Press: 03 April 2017
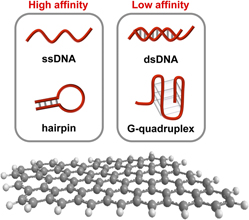
Single-stranded DNA molecules capable of molecular recognition and catalysis can now be routinely generated via the technique of in vitro selection. When coupled with adequate signal transduction modes, these synthetic functional DNA species represent a potential paradigm shift in the research and development of biosensors to meet the challenges of our rapidly changing world. Coupling functional DNA molecules with graphene materials for the design of optical biosensors has become an exciting research area in recent years, mostly because graphene materials are not only excellent quenchers of fluorescence, but they also display considerably different affinities for free and ligand-bound functional DNA molecules. We will discuss notable progress in this area in this mini-review by highlighting representative studies.
Contributing Editor: Venkatesan Renugopalakrishnan