Introduction
Metaxonchium Coomans & Nair, Reference Coomans and Nair1975 is an interesting, almost cosmopolitan and diverse dorylaimid (Nematoda, order Dorylaimida) genus. Originally proposed as a subgenus of the genus Axonchium Cobb, Reference Cobb1920, it was raised to generic rank by Andrássy (Reference Andrássy and Mahunka1996). It currently includes 30 valid species plus one species inquirenda, which have been recorded in at least 28 countries or archipelagos, most of them in Holarctic territories (Peña-Santiago, Reference Peña-Santiago2021).
Iranian nematode fauna of belondirid (family Belondiridae) taxa remain poorly known, and Metaxonchium representatives are not an exception. Thus, only two species, namely Metaxonchium bihariense (Popovici, Reference Popovici1990) Andrássy, Reference Andrássy and Mahunka1996 and Metaxonchium persicum Peña-Santiago, Niknam, Álvarez-Ortega & Jabbari, Reference Peña-Santiago, Niknam, Álvarez-Ortega and Jabbari2014, were recorded in the country throughout the last decade (Peña-Santiago et al., Reference Peña-Santiago, Niknam, Álvarez-Ortega and Jabbari2014; Vazifeh et al., Reference Vazifeh, Niknam and Jabbari2018; Jabbari et al., Reference Jabbari, Niknam, Fallahi, Zahedi Asl and Nikdel2019; Kazemi & Niknam, Reference Kazemi and Niknam2019).
A nematological survey conducted to explore nematode diversity of Iran yielded a population of a comparatively large-sized Metaxonchium species. Its morphological and molecular characterization revealed that it belonged to an undescribed species and provided new insights into the phylogeny of the genus. The results of this study are presented in the following.
Material and methods
Sampling and processing of nematodes
Soil samples were collected from natural habitats in Darram village, Tarom County, Zanjan Province, northwestern Iran. Nematodes were extracted using the tray method (Whitehead & Hemning, Reference Whitehead and Hemning1965), handpicked under a stereomicroscope, killed by adding hot FPG (4:1:1, formaldehyde: propionic acid: glycerine) solution, transferred to anhydrous glycerine according to De Grisse (Reference De Grisse1969) and mounted on permanent glass slides to allow handling and observation.
Morphological and morphometrical study
A total of 11 females and ten males, in good state of preservation, were available. Nematodes were measured and photographed using an Eclipse 80i microscope (Nikon, Tokyo, Japan) equipped with differential interference contrast optics, a drawing tube (camera lucida) and a DS digital camera. Morphometrics include Demanian indices and other measurements and ratios, some of them presented in a separate table, meanwhile the remaining measurements and ratios form part of the literal description of species.
Scanning electron microscopy (SEM)
Specimens preserved in glycerine were selected for their observation with a SEM according to Abolafia (Reference Abolafia2015). The nematodes were hydrated in distilled water, dehydrated in a graded ethanol–acetone series, critical point dried, coated with gold, and observed with a Zeiss Merlin microscope (5 kV) (Zeiss, Oberkochen, Germany).
DNA extraction, polymerase chain reaction (PCR) and sequencing
Following morphological confirmation, five fresh specimens of Metaxonchium magnum sp. n., were selected for DNA extraction. Each specimen was transferred to an Eppendorf tube containing 20 μl of nuclease-free water. DNA was extracted using the modified Chelex method (Rashidifard et al., Reference Rashidifard, Marais, Daneel, Mienie and Fourie2019) by adding 25 Chelex (5% w/v) and 5 μl of proteinase K – the tubes were incubated at 56°C for 2 h followed by 95°C for 10 min and then stored at −18°C until further processing. Five μl of each extracted DNA was added to the PCR mixture in a 0.2 ml Eppendorf tube containing: 15 μl 2× Master mix (Ampliqon, Denmark); 1 μl of each primer (10 pMol/μl); and 8 μl double-distilled water, to a final volume of 30 μl. The D2–D3 expansion segments of 28S rDNA were amplified using forward D2A (5′–ACAAGTACCGTGAGGGAAAGTTG–3′) and reverse D3B (5′–TCGGAAGGAACCAGCTACTA–3′) primers (Nunn, Reference Nunn1992). PCR reactions were carried out in a DNA thermal cycler (Hybaid, Ashford, Middlesex, UK). The PCR programme was followed as reported by Rashidifard et al. (Reference Rashidifard, Bello, Fourie, Coyne and Peña-Santiago2020). The quality of PCR were checked by electrophoresis of 4 μl of the PCR reactions in 1% agarose gel containing SYBR Green I. Products were visualized and photographed under ultraviolet light. The length and concentration of each PCR product was measured by comparison with a low DNA mass ladder (Invitrogen, Carlsbad, CA). The PCR products were purified and sequenced directly for both strands using the same primers with an ABI 3730XL sequencer (Bioneer, Seoul, South Korea). The newly obtained sequences of the 28S region were submitted to the GenBank database under accession numbers OQ099690-4.
Phylogenetic analyses
For phylogenetic relationships, analysis was based on 28S rDNA fragments containing gene coding for 28S rRNA. The newly obtained sequences were manually edited using Chromas 2.6.6 (Technelysium, Queensland, Australia) and aligned with another 28S rDNA sequences available in GenBank using ClustalW alignment tool implemented in the MEGA7 (Kumar et al., Reference Kumar, Stecher and Tamura2016). Poorly aligned regions at extremes were removed from the alignments using MEGA7. The best-fit model of nucleotide substitution used for the phylogenetic analysis was statistically selected using jModelTest 2.1.10 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012). The phylogenetic tree was generated with the Bayesian inference method using MrBayes 3.2.6 (Ronquist et al., Reference Ronquist, Teslenko and van der Mark2012). Mononchus truncatus Bastian, Reference Bastian1865 (AY593064), was chosen as outgroup. The analysis under the generalized time reversible and invariant sites and gamma distribution (GTR + I + G) model was initiated with a random starting tree and run with the Markov chain Monte Carlo (Larget & Simon, Reference Larget and Simon1999) for 1 × 106 generations. The tree was visualized and saved with FigTree 1.4.4 (Rambaut, Reference Rambaut2018).
Results
M. magnum sp. n. (figs 1–4, morphometrics in table 1)

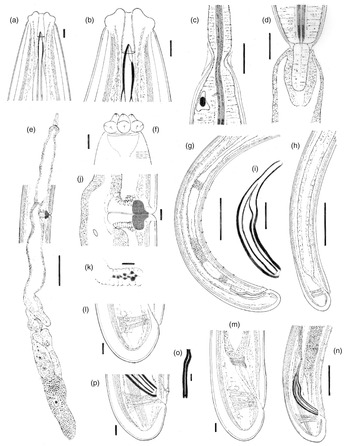
Fig. 1. Metaxonchium magnum sp. n. from Iran (drawings): (a, b) anterior body region, in lateral median view; (c) isthmus-like narrowing between the two pharyngeal sections; (d) pharyngo-intestinal junction; (e) female, genital system; (f) anterior body region, in surface lateral view; (g) male, posterior body region; (h) female, posterior body region; (i) spicule; (j) vagina region; (k) uterine Z-like organ; (l) female, caudal region; (m) female, rectum and caudal region; (n) male, spicule and caudal region; (o) lateral guiding piece; and (p) male, caudal region. Scale bars: (a), (b), (f), (o) 5 μm; (c), (d), (i) 20 μm; (e), (g), (h) 100 μm; (j)–(m), (p) 10 μm; (n) 50 μm.

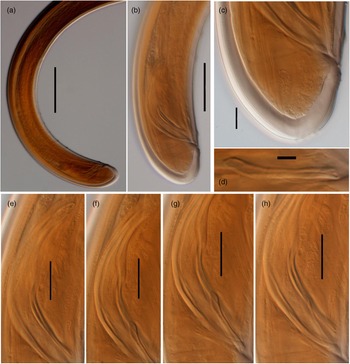
Fig. 2. Light microscopy micrographs of Metaxonchium magnum sp. n. from Iran (female): (a, b) anterior body region, in lateral median view; (c) posterior genital branch; (d) pharyngo-intestinal junction; (e) isthmus-like narrowing between the two pharyngeal sections; (f) anterior body region, in surface lateral view; (g) uterine Z-like organ; (h, j) vagina region; (i) posterior body region; (j) Rectum and caudal region; and (l, m) caudal region. Scale bars: (a), (b), (f) 5 μm; (c), (i) 100 μm; (d), (e) 20 μm; (g), (h), (j), (l), (m) 10 μm; (k) 50 μm.

Fig. 3. Light microscopy micrographs of Metaxonchium magnum sp. n. from Iran (male). (a, b) posterior body region; (c) caudal region; (d) lateral guiding piece; (e–h) spicule. Scale bars: (a) 100 μm; (b) 50 μm; (c) 10 μm; (d) 5 μm; (e)–(h) 20 μm.

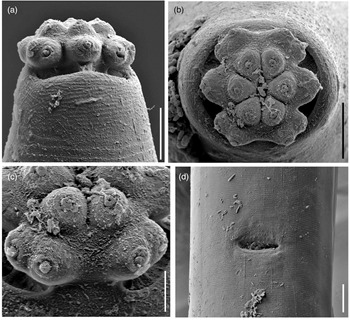
Fig. 4. Scanning electron microscopy micrographs of Metaxonchium magnum sp. n. from Iran. (a) anterior region, in lateral view; (b) same, in face view; (c) detail of subventral lips; and (d) vulva, in ventral view. Scale bars: (a), (b) 5 μm; (c) 2 μm; (d) 10 μm.
Table 1. Morphometrics of Metaxonchium magnum sp. n. Measurements in μm except L in mm and in the form: mean ± standard deviation (range).

♀, female; ♂, male.
Adult: Slender to very slender nematodes (a = 42–55) of large size, 3.62–4.65 mm long. Body cylindrical, visibly tapering towards the anterior end, less so towards the posterior end as the caudal region is short and rounded. Habitus curved ventrad upon fixation, C-shaped or G-shaped. Cuticle two-layered, with very fine transverse striation only perceptible under SEM, 3.5–5 μm thick in anterior region, 5–7 μm at mid-body and 11–13 μm on dorsal side of female tail and 8–11 μm on dorsal side of male tail, consisting of inner layer thicker than the outer one, especially obvious at caudal region where it bears distinct radial striation. Cervical lacunae conspicuous. Lateral chord appreciably narrow, 7–9 μm or up to one-tenth (9–10%) of mid-body diameter in females and 3–6 μm or 4–6% of mid-body diameter in males, of granular nature and lacking any other differentiation. Lip region typically cap-like, offset by constriction, 2.3–3.0 times as broad as high or one-sixth to one-fifth (15–20%) of body diameter at neck base; lips variably distinct (as seen under SEM), outer portions of the lateral lips separated from the subdorsal and the subventral ones by deep incisures meanwhile the subventral and the subdorsal appear more amalgamated, inner (perioral) region of lips forming distinct, somewhat triangular lobes that are offset between them by the existence of six radial incisures; and labial and cephalic papillae button-like, with a pore at their centre, slightly interfering with the labial contour. Amphid fovea cup-like, its aperture 10–12 μm wide, occupying more than three-quarters (76–84%) of lip region diameter. Cheilostom a truncate cone, with comparatively thin walls. Odontostyle small, somewhat fusiform, almost equal to (0.8–1.2 times) than lip region diameter, 3.8–5.2 times as long as wide and 0.31–0.44% of body length; aperture 4.0–5.5 μm long, occupying one-fifth to two-fifths (26–42%) of total length. Guiding ring thin, simple, but visibly refractive, at 12–14 μm or 0.8–1.1 times the lip region diameter from the anterior end. Odontophore rod-like, 1.3–1.6 times the odontostyle. Pharynx consisting of a slender and weakly muscular anterior portion, which is separated from the basal expansion by a short isthmus-like narrowing; basal expansion showing a remarkable sexual dimorphism: 17.5–23.0 times as long as broad, 9.5–12.2 times longer than body diameter at neck base, and occupying 73–77% of the total neck length in females, and 13.4–17.3 times as long as broad, 7.2–10.7 times longer than body diameter at neck base, and occupying 66–72% of the total neck length in males; and a very distinct spiral muscular sheath, with nearly straight muscular bands, envelopes the whole basal expansion. Cardia tongue-like, 33–56 × 12–17 μm, surrounded by intestinal tissue. Caudal region short and rounded, almost hemispherical in females and slightly more conoid in males; and caudal pores two pairs at the posterior half of tail, one lateral, another sublateral.
Female: Genital system mono-opistho-ovarian, didelphic. Anterior branch 260–520 μm long or 7–14% of body length, and consisting of a long uterine sac often not containing sperm cells, a small but distinct sphincter, and a solid terminal mass 39–70 μm long, probably representing remains of oviduct and/or ovary. Posterior branch very long, 700–950 μm, often more or less convoluted. Reflexed ovary large, 150–432 μm long, with oocytes arranged first in several rows and then in a single row. Oviduct joining the ovary subterminally, 133–304 μm long or 1.7–3.2 body diameters, and consisting of a tubular part made of prismatic cells and a well-developed pars dilatata with distinct lumen and occasionally containing sperm cells. A strong sphincter separates oviduct and uterus. Uterus 538–600 μm or 5.2–6.9 times the body diameter long, always convoluted, tripartite, that is, consisting of a large proximal section with wide lumen and visibly (and unusually) muscular walls, a convoluted intermediate region with narrow lumen and a Z-like differentiation bearing granular elements at its distal portion, and a large spherical distal pars dilatata; and sperm cells are often present and abundant, especially at proximal and distal parts of uterus. Uterine eggs 146–180 × 46–68 μm. Vagina 43–56 μm long, extending inwards to one-half (49–52%) of the corresponding body diameter: pars proximalis longer than wide, 23–31 × 14–17 μm, with nearly straight walls and surrounded by strong circular musculature; pars refringens (in lateral view) wider than pars proximalis, consisting of two close together trapezoidal to drop-shaped pieces measuring 10–13 × 14.5–15 μm and with a combined width of 22–29 μm; and pars distalis 5.5–8.5 μm long. Vulva an equatorial transverse slit, approximately 15 μm long. Prerectum 5.1–7.7, rectum 1.0–1.2 anal body diameters long.
Male: Prerectum 7.1–9.2, cloaca 1.4–1.5 body diameters long. Genital system diorchic, with opposite testes. In addition to the ad-cloacal pair, situated at 12–15 μm from cloacal aperture, there is a series of 10–13 spaced (20–32 μm apart) ventromedian supplements, the posterior most of which is situated at 52–77 μm from the ad-cloacal pair, with hiatus. Spicules dorylaimid, 6.2–7.2 times as long as broad, 1.6–1.9 times the corresponding body diameter: head occupying 25–35% of the total length, 4.3–6.4 times as long as wide, with its dorsal side visibly longer than the ventral one; median piece occupying one-half (44–54%) of maximum spicule wide; posterior end 6–7.5 μm wide; ventral hump located at 39–50 μm from the anterior end; and curvature 115–126°. Lateral guiding pieces 29–32 μm long, 10–13 times as long as wide, slightly curved ventrad and with bifid end.
Molecular characterization
Five sequences for the 28S rDNA, one with 759 base pairs (bp) (OQ099690), one with 767 bp (OQ099691), one with 742 bp (OQ099692) and two with 778 bp (OQ099693-4) were obtained for M. magnum sp. n., all of them with identical sequences in a fragment in common with 707 bp.
The Basic Local Alignment Search Tool using the longest newly generated D2–D3 sequence revealed it has 89.05% identity with the same genomic region of Metaxonchium stenospiculum (accession number MH167346-8), and 84.47% identity with the same genomic region of Metaxonchium toroense (MG MG018767-9) in a fragment in common with 703 bp. The other high identities belonged to species of other genera such as Axonchoides smokyensis (JX885739, 88.58% identity in fragment in common with 718 bp) and Syncheilaxonchium nairi (MF325350-1, 95.00% identity in a fragment in common with 280 bp).
Diagnosis
The new species is characterized by its 3.62–4.65 mm long body, lip region cap-like and offset by constriction and 13–16 μm wide, odontostyle fusiform and 14–17 μm long, neck 1016–1359 μm long, both parts of the pharynx separated by a short isthmus-like narrowing, pharyngeal expansion occupying 73–77% of total neck length in females and 66–72% in males, female genital system mono-opistho-ovarian, didelphic, anterior genital branch reduced to a large uterine sac and a small terminal mass occupying 7–14% of body length, posterior uterus long and tripartite with a Z-like differentiation, V = 50–52, caudal region short and rounded (24–41 μm, c = 99–161, c′ = 0.5–0.7), spicules 90–105 μm long and 10–13 spaced ventromedian supplements with hiatus.
Relationships
Morphologically, and in its general appearance and its uterus containing sclerotized elements, the new species resembles M. persicum Peña-Santiago, Niknam, Álvarez-Ortega & Jabbari, Reference Peña-Santiago, Niknam, Álvarez-Ortega and Jabbari2014, another Iranian species, but it can be easily distinguished from this by its larger general size (body 3.62–4.65 vs. 2.46–3.12 mm long), wider lip region (13–16 vs. 8–11 μm), longer odontostyle (14–17 vs. 10–12 μm long), absence (vs. presence) of a spindle-like thickening at the anterior portion of pharynx, uterus with a Z-like differentiation (vs. spine-like elements or apophyses present throughout its intermediate section), more anterior vulva (V = 50–52 vs. V = 53–57), and hiatus present (vs. at least two ventromedian supplements situated within the spicule range). Among the species of the genus lacking sclerotized elements in the uterus, M. magnum sp. n. resembles a reduced group of large-sized species, their body being very often over 3.0 mm long, namely Metaxonchium coxi (Yeates, Reference Yeates1979) Peña-Santiago Niknam, Álvarez-Ortega & Jabbari, Reference Peña-Santiago, Niknam, Álvarez-Ortega and Jabbari2014, Metaxonchium crassum (Thorne, Reference Thorne1939) Andrássy, Reference Andrássy and Mahunka1996, Metaxonchium gigas (Thorne, Reference Thorne1939) Andrássy, Reference Andrássy and Mahunka1996 and M. toroense Varela-Benavides & Peña-Santiago, Reference Varela-Benavides and Peña-Santiago2019. It differs from M. coxi, a poorly described species only known from New Zealand (Yeates, Reference Yeates1979; Yeates & Van Etteger, Reference Yeates and van Etteger1991), in its wider lip region (13.0–14.5 μm in the new species vs. approximately 11 μm in M. coxi, calculated from Yeates’ original illustrations), longer odontostyle (15–17 vs. approximately 12 μm), female tail almost hemispheroid (vs. convex conoid) and shorter (24–39 vs. 38–50 μm, c = 99–161 vs. 71–106, c′ = 0.7–0.8 vs. 0.5–0.7), and males as frequent as females (vs. monosexual, with only females known, n = 25). From M. crassum, a North American taxon (Thorne, Reference Thorne1939; Hechler, Reference Hechler1969; Nair, Reference Nair1973), in its shorter odontostyle (14–17 vs. 21–23 μm), more anterior vulva (V = 50–52 vs. V = 56–58), and in being a bisexual species with both females and males equally frequent (vs. monosexual, with only females known). From M. gigas, also a North American species (Thorne, Reference Thorne1939; Hechler, Reference Hechler1969; Nair, Reference Nair1973), in its shorter odontostyle (14–17 vs. 17–19 μm), and males with less ventromedian supplements (10–13 vs. 17) differently arranged (hiatus present vs. absent). From M. toroense, a Mesoamerican taxon, in its larger general size (body 3.62–4.65 vs. 2.44–3.31 mm long), much longer anterior genital branch (7–14 vs. 1–2% of body length) and males as frequent as females (vs. males absent).
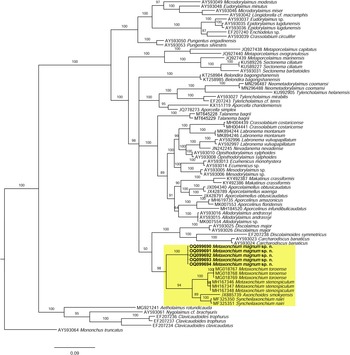
As derived from the molecular analysis of D2–D3 28S rRNA, whose results are presented in the tree of fig. 5, the new sequences herein obtained (OQ099690-4) cluster in a highly supported (posterior probability (PP) = 98%) clade with sequences of other Metaxonchium species, namely M. stenospiculum Varela-Benavides & Peña-Santiago, Reference Varela-Benavides and Peña-Santiago2018 and M. toroense, as well sequences of other two belondirid genera and species, A. smokyensis Peña-Santiago, Abolafia, Álvarez-Ortega, Ye & Robbins, Reference Peña-Santiago, Abolafia, Álvarez-Ortega, Ye and Robbins2013 and S. nairi (Altherr, Reference Altherr1974) Coomans & Nair, Reference Coomans and Nair1975, so confirming previous findings (Varela-Benavides & Peña-Santiago, Reference Varela-Benavides and Peña-Santiago2018, Reference Varela-Benavides and Peña-Santiago2019). Nevertheless, the present results reveal that sequences of the new species form a subclade separated from that constituted by the remaining sequences ((M. stenospiculum + M. toroense) + (Syncheilaxonchium Coomans & Nair, Reference Coomans and Nair1975 + Axonchoides Thorne, Reference Thorne1967)), which shows slightly lower support (PP = 94). This suggests that Metaxonchium might be a non-monophyletic taxon and that its evolutionary relationships with other rounded-tailed belondirid genera should be a matter of further analysis.

Fig. 5. Bayesian inference tree from the newly sequenced Metaxonchium magnum sp. n. based on sequences of the 28S rDNA region. Bayesian posterior probabilities (%) are given for each clade. Scale bar shows the number of substitutions per site.
Type locality and habitat
Northwestern Iran, Zanjan Province, Tarom County, Darram village, (Global Positioning System coordinates: 37° 07′ 24.475″ N, 48° 84′ 61.375″ E; altitude: 650 m above sea level), where the new species was recovered from soil samples around the feeder roots of a walnut tree (Juglans regia L.) located in the proximity of a small pond, collected on 20 September 2021.
Type material
Female holotype, one female paratype and two male paratypes deposited at the Nematode Collection of the Departamento de Biología Animal, Biología Vegetal y Ecología, Universidad de Jaén, Spain. Seven female paratypes and six male paratypes deposited at the nematode collection of the Faculty of Agriculture, University of Zanjan, Zanjan, Iran. One female paratype and one male paratype deposited at the United States Department of Agriculture Nematode Collection, Beltsville, Maryland, USA.
Etymology
The specific epithet, magnum, is a Latin term meaning large, and refers to the comparatively large size of the new species.
Acknowledgements
For assistance to obtain scanning electron microscopy pictures of the new species herein described, the Spanish authors are grateful to Amparo Martínez-Morales and the equipment provided by ‘Centro de Instrumentación Científico-Técnica (CICT)’, University of Jaén, Spain.
Financial support
University of Jaén, Spain, through the research programme ‘PAIUJA 20121/2022: EI_RNM02_2021’.
Conflicts of interest
None.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.