Introduction
Capsalid monogeneans belonging to the genus Encotyllabe Diesing, Reference Diesing1850 are ectoparasitic Monopisthocotylea. They are commonly found on various parts of marine fish, such as the gills, body surface, mouth and pharyngeal toothpads of marine fish. Species of this genus are recognized by the elliptical ventral suckers surrounded by a wide membrane and a pedunculated aseptate haptor equipped with two pairs of substantial hooks and several marginal hooklets (Price Reference Price1939).
Encotyllabe spp. are known to inhabit a wide range of waters, encompassing temperate, subtropical and tropical regions (Lebedev Reference Lebedev1967). They have a broad host spectrum (Table 1), infecting various teleost fish families (Sepúlveda et al. Reference Sepúlveda, González and Oliva2014). Currently, there are 24 species attributed to this genus (WoRMS 2023), however, the validity of several of these species has been questioned due to the incomplete and brief original descriptions (Taborda et al. Reference Taborda, Sepulveda, Luque, Escribano and Oliva2023), the unavailability or poor conditions of some type specimens (Sepúlveda et al. Reference Sepúlveda, González and Oliva2014) and the lack of detailed redescriptions. Additionally, seven of the total recognized species were initially described based on only one or two specimens, which hinders the ability to understand intraspecific variability (Sepúlveda et al. Reference Sepúlveda, González and Oliva2014). Occasionally, minor morphoanatomical differences have been deemed sufficient grounds for the creation of new species (Khalil and Abdul-Salam Reference Khalil and Abdul-Salam1988; Martorell Reference Martorell2004).
Table 1. The distribution of Encotyllabe spp. according to the systematics of host groups, reproduced from Lebedev (Reference Lebedev1967), updated. Note that E. latridis mentioned by Lebedev (Reference Lebedev1967) was omitted because it is currently included in Mediavagina Lawler and Hargis, 1968. Encotyllabe masu Ishii and Sawada, Reference Ishii and Sawada1938; E. monticelli Perez Vigueras, 1940; E. pricei Koratha, 1955 and E. punctatai Gupta et Krishna, 1980 as species inquirendae because these species were poorly described, based on only one or two specimens.

1 was originally described in an unpublished thesis and never published according to the rules of the International Code for Zoological Nomenclature; it is not considered valid (see Taborda et al. (Reference Taborda, Sepulveda, Luque, Escribano and Oliva2023)).
2 Yamaguti (Reference Yamaguti1934) noted that both E. pagelli and E. pagrosomi are parasitic in the sparid Pagrus auratus and probably more closely related to E. spari, but because the original papers of these species were inaccessible to him, he noted that his species should be looked upon as a provisional one.
Only three Encotyllabe species occur in the Mediterranean: Encotyllabe paronae Monticelli, Reference Monticelli1907, described from the East Atlantic peacock wrasse Symphodus tinca Linnaeus, 1758 (Labridae); Encotyllabe nordmanni Diesing, Reference Diesing1850, described from two Bramidae, the Mediterranean pomfret Brama mediterranea (Bonnaterre, 1788) and the Atlantic pomfret Brama brama (Bonnaterre 1788) and a Pomacentridae Chromis (Linnaeus 1758). Two species are known from sparid fishes: Encotyllabe pagelli Van Beneden and Hesse, Reference Van Beneden and Hesse1863, from the blackspot seabream Pagellus centrodontus (Delaroche 1809) collected off Brest, France, and E. vallei Monticelli, Reference Monticelli1907, described from the gilt-head bream Sparus aurata Linnaeus, 1758 (Sparidae Rafinesque 1818) off Trieste, Italy. Encotyllabe vallei had been rarely mentioned, and apart from Radujkovic and Euzet (Reference Radujkovic, Euzet, Radujkovic and Raibaut1989), the previous accounts provided scant or no morphometric data and lacked illustrations. Consequently, the brief original description has made it difficult to discern differences from other species (Taborda et al. Reference Taborda, Sepulveda, Luque, Escribano and Oliva2023). As a result, assessing its similarities to and differences from other Encotyllabe species remains challenging due to the absence of morphometric data which led to its recent classification as species inquirendae (Taborda et al. Reference Taborda, Sepulveda, Luque, Escribano and Oliva2023).
In this study, we provide an illustrated redescription of E. vallei based on newly collected specimens from the type-host S. aurata collected from the western Mediterranean, Algeria. We also provide cox1 and 28S sequences and discuss the host specificity of E. vallei.
Material and methods
During the years 2020–2021, 115 specimens of S. aurata were collected from off Dellys (36°54′ 48″N, 3°54′ 51″E), Zemmouri El Bahri (36°48′ 4.58″N, 3°34′ 7.01″E), Cap Djinet (36°52′ 37″ N, 3°43′ 23″E), Bouharoun (36′370 24″ N, 2°39′ 17″ E) and Cherchell (36°36′ 31″ N, 2°11′ 50″ E) off the Algerian coast. Fish specimens were purchased dead from fishermen, transferred to the laboratory shortly after capture, identified using keys (Fischer et al. Reference Fischer, Bauchot and Schneider1987; Kullander and Delling Reference Kullander, Nyman, Jilg and Delling2012) and examined fresh on the day of purchase.
Gills and pharyngeal tooth pads were detached, placed in individual Petri dishes containing saline solution following Lablack et al. (Reference Lablack, Rima, Georgieva, Marzoug and Kostadinova2022) and carefully examined for monogeneans under a stereomicroscope (Carl Zeiss Microimaging GmbH 37081 Göttingen, Germany). Monogeneans were removed from gills using fine dissection needles, heat-killed and preserved in molecular biology-grade ethanol and 70% ethanol for molecular and morphological analyses, respectively. We followed the terminology as defined by Combes (Reference Combes2003) to describe the host specificity of a parasite in relation to the relatedness of host species: Oioxenic is employed for parasites that exploit a single host species; the parasite is denoted as stenoxenic if it exploits a range of phylogenetically related species and euryxenic if it exploits a range of mutually unrelated species.
Morphological methods
For morphological study, monogeneans were stained with acetic carmine, dehydrated in graded ethanol series (70%, 95% and 100%), cleared in clove oil and mounted in Canada balsam using a Wild Heerbrugg stereo microscope. Voucher material was deposited at the Swedish Natural History Museum, Stockholm, Sweden, under registration numbers SMNH-208363–208373, SMNH-218760–218780. Drawings were made with the help of a Nikon Eclipse 80i microscope equipped with differential interference contrast (DIC) and a drawing tube (Department of Zoology, Swedish Museum of Natural History). Drawings were scanned and redrawn on a computer with Adobe Illustrator 2023. Measurements are in micrometers and indicated as the range followed by the mean.
Molecular methods
Three monogeneans were selected for DNA extraction. For three S. aurata, one monogenean was extracted. A tissue sample from the gill of the fish was taken, preserved in absolute ethanol and deposited as a voucher in the SMNH. For the monogenean, a small lateral part of the body above the haptor was separated with a scalpel following previous works on the barcoding of Monogenea (Ayadi et al. Reference Ayadi, Tazerouti, Gastineau and Justine2022; Azizi et al. Reference Azizi, Bouguerche, Santoro, Gey, Tazerouti, Justine and Bahri2021; Bouguerche et al. Reference Bouguerche, Gey, Justine and Tazerouti2019a, Reference Bouguerche, Gey, Justine and Tazerouti2019b; Bouguerche et al. Reference Bouguerche, Tazerouti, Gey and Justine2019; Bouguerche et al. Reference Bouguerche, Tazerouti, Gey and Justine2020; Bouguerche et al. Reference Bouguerche, Tazerouti, Gey and Justine2021; Lablack et al. Reference Lablack, Rima, Georgieva, Marzoug and Kostadinova2022) and submitted to molecular analysis, and the rest of the body was mounted on a slide as a voucher for drawing and deposition in a museum collection (Figure 1A). This enables the morphological assessment of sequenced monogeneans. Slides of monogeneans used for molecular work were deposited in the SMNH under registration numbers SMNH-208 363, SMNH-208 364 and SMNH-208 365.

Figure 1. Encotyllabe vallei (Monticelli, Reference Monticelli1907) ex S. aurata. A, Hologenophores SMNH-208 365 (GenBank OR148273); B, voucher, whole body, SMNH-208 366; C, Large hamulus; D, small hamulus.
Molecular barcoding of Monogenea
For cox1, total genomic DNA was isolated using a QIAmp DNA Micro Kit (Qiagen). The specific primers JB3 (=COIASmit1) (forward 50–TTTTTTGGGCATCCTGAGGTTTAT–30) and JB4.5 (=COI-ASmit2) (reverse 50–TAAAGAAAGAACATAATGAAAATG–30) were used to amplify a fragment of 402 bp of the cox1 gene (Bowles et al. Reference Bowles, Blair and McManus1995; Littlewood et al. Reference Littlewood, Rohde and Clough1997). PCR reactions were performed in 20 μl of a mixture containing 1 ng of DNA, 1 CoralLoad PCR buffer, 3 mM MgCl2, 0.25 mM dNTP, 0.15 μM of each primer and 0.5 units of Taq DNA polymerase (Qiagen). Thermocycles consisted of an initial denaturation step at 94°C for 2 min, followed by 37 cycles of denaturation at 94°C for 30 s, annealing at 48°C for 40 s and extension at 72°C for 50 s. The final extension was conducted at 72°C for 5 min. PCR products were purified (Ampure XP Kit, Beckman Coulter) and sequenced in both directions on a 3730 l DNA Analyzer 96-capillary sequencer (Applied Biosystems, Foster City, CA, USA). We used CodonCode Aligner version 3.7.1 software (Codon Code Corporation, Dedham, MA, USA) to edit sequences and compare them to the GenBank database content with BLAST and deposited them in GenBank under accession numbers OR148271, OR148272, OR148273.
For 28S, DNA was extracted using a QiaAmp DNA Micro Kit (Qiagen). A 28S rDNA fragment of 884 bp was amplified using the universal primers C10 (50–ACCCGCTGAATTTAAGCAT–30) and D2 (30–TCCGTGTTTCAAGACGG–50) (Hassouna et al. Reference Hassouna, Michot and Bachellerie1984). PCR reactions were performed in a final volume of 20 mL, containing: 1 ng of DNA, 16 CoralLoad PCR buffer, 3 mM MgCl2, 66 mM of each dNTP, 0.15 mM of each primer and 0.5 units of Taq DNA polymerase (Qiagen). Thermocycles consisted of an initial denaturation step at 94°C for 1 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s and extension at 72°C for 1 min. The final extension was conducted at 72°C for 7 min. PCR products were visualized on a 1.5% agarose gel, purified and directly sequenced in both directions on a 3730xl DNA Analyzer 96-capillary sequencer (Applied Biosystems) at Eurofins Genomics. Sequences were edited and assembled using CodonCode Aligner software (CodonCode Corporation, Dedham, MA, USA) and compared to the GenBank database content with BLAST. Sequences from three individual monogeneans were obtained and were found to be identical; they were deposited in GenBank under accession numbers OR149163, OR187608.
Molecular analysis
Trees were constructed using our newly generated sequences and those of closely related taxa available in GenBank. For 28S, we used almost all closely related sequences available in GenBank (Table 2), except for those that were too short and/or did not align well. A sequence of a monocotylid Holocephalocotyle monstrosae Derouiche, Neifar, Gey, Justine and Tazerouti, Reference Derouiche, Neifar, Gey, Justine and Tazerouti2019 (Derouiche et al. Reference Derouiche, Neifar, Gey, Justine and Tazerouti2019) was used as an outgroup. The 28S dataset included 50 nucleotide sequences. Both extremities of the sequences were trimmed to obtain a clean matrix. After estimating the best model with MEGA7 (Kumar et al. Reference Kumar, Stecher and Tamura2016), the tree was inferred using the maximum likelihood (ML) method based on the general time reversible model (GTR) with gamma distributed with invariant sites (G+I).
Table 2. Sequences used in the molecular analysis of 28S sequences of monogeneans.

*, new sequences.
For cox1, the newly generated sequences of E. vallei were aligned with 25 species of Encotyllabe recovered after BLAST (Table 3). Two sequences of Empruthotrema aoneken Irigoitia, Braicovich, Rossin and Timi in Irigoitia, Braicovich, Rossin, Canel, Levy, Farber and Timi, Reference Irigoitia, Braicovich, Rossin, Canel, Levy, Farber and Timi2019 (MN190708, MN190709 (Irigoitia et al. Reference Irigoitia, Braicovich, Rossin, Canel, Levy, Farber and Timi2019)) were used as an outgroup. The trimmed matrix included 261 positions in the dataset. The tree was inferred using the ML method based on the Hasegawa–Kishino–Yano model (HKY) with gamma distributed (G).
Table 3. Sequences used in the molecular analysis of cox1 sequences of monogeneans.

*, new sequences. numbers SNHM 208 363-208 373, SMNH 218 760-218 780.
All trees were constructed in MEGA7, with 500 replications for the ML trees. The neighbour-joining (NJ) method was also used for comparison in MEGA7, with 2000 bootstraps computed for cox1 and 28S from the same datasets. Distances (p-distances and Kimura two parameter) were computed from the same datasets with MEGA7(Kumar et al. Reference Kumar, Stecher and Tamura2016).
Results
Molecular characterization of monogeneans
For the 28S gene, molecular data were generated for three specimens of E. vallei collected from S. aurata off Algeria, Western Mediterranean (OR148271, OR148272, OR148273). The alignment of the new 28S sequence data with that of related taxa, primarily Encotyllabinae Monticelli, 1892, Benedeniinae Johnston and Capsalinae Baird, 1853, yielded 1288 characters. The final dataset used for phylogenetic analysis included 846 characters. The phylogenetic analysis using both ML and NJ methods produced phylograms with identical topologies (Figure 2). In these phylograms, each of the four capsalid subfamilies represented a distinct and well-supported clade. Within Encotyllabe, three subclades were identified: the first included Encotyllabe sp1. collected from P. bogaraveo, while the second included E. vallei from S. aurata and D. vulgaris. The third subclade within Encotyllabe included the following species: Encotyllabe sp. from Pagrus pagrus, Encotyllabe sp. from Orthopristis ruber, E. caballeroi, Encotyllabe cheilodactyli, Encotyllabe chironemi, E. cf. spari and Encotyllabe antofagastensis.

Figure 2. Maximum likelihood tree based on an analysis of 28S sequence data for Capsalidae. Bootstrap percentages (500 replicates) are indicated next to or below the branches (only values >70% are shown). There was a total of 846 positions in the final dataset. The NJ tree (p-distance method) had a similar topology and is not presented.
There were no intraspecific variations within our newly generated sequences of E. vallei. Additionally, all sequences of E. vallei from the Western Mediterranean (off Algeria) were identical, irrespective of host. Overall, the genetic distance between the newly generated sequences and those of Capsalidae ranged from 1% to 33%, while the divergence among Encotyllabe spp. ranged from 1% to 2%. The closest sequences to our newly generated sequences of E. vallei were those of E. chironemi from Chironemus marmoratus off Australia, Encotyllabe cf. spari from O. ruber off Brazil, Encotyllabe sp. from P. pagrus, and from O. ruber off Brazil, and E. antofagastensis from Anisotremus scapularis off Chile (1% interspecific variation).
The newly generated cox1 sequences (OR148271, OR148273 and OR148273, respectively) were 433, 416 and 432 bp long. Deleted ambiguously aligned characters resulted in a final dataset of 261 characters for phylogenetic analysis. The ML and NJ analyses also have identical topologies and only the ML tree is shown (Figure 3). In the cox1 dataset, the same clades (when cox1 and 28S sequences are both available for a given species) were represented with much greater levels of difference between them. All sequences of E. vallei from Algeria clustered in a well-supported clade. Similarly, Encotyllabe sp. and Encotyllabe sp. 2 from O. ruber and A. scapularis, respectively, formed well-supported monophyla.

Figure 3. Maximum likelihood tree based on an analysis of cox1 sequence data for Encotyllabe spp. Bootstrap percentages (500 replicates) are indicated next to or below the branches (only values > 70% are shown). There was a total of 261 positions in the final dataset. The NJ tree (p-distance method) had a similar topology and was not shown.
The replicate cox1 sequences of E. vallei generated in the present study differed between them by 0% to 1% (intraspecific variations). All genotypes of E. vallei from Algeria from different hosts differed by 1% to 2% divergence in P-distances.
Morphology of E. vallei Monticelli, Reference Monticelli 1907 (Figures 1 and 4 )
Type-host: S. aurata (Linnaeus), the gilt-head (sea) bream (Sparidae).
Additional hosts: Dentex sp., D. dentex (Linnaeus.), Diplodus puntazzo (Walbaum) and Mullus surmuletus Linnaeus.
Type-locality: Trieste, Italy (Monticelli, Reference Monticelli1907).

Figure 4. Encotyllabe vallei (Monticelli, Reference Monticelli1907) ex S. aurata, Detail of the reproductive organs in the region of the vagina (SMNH-208 367).
Additional localities: Montenegro, Spain. Off Bouharoun, off Dellys (36°54′ 48″N, 3°54′ 51″E), Zemmouri El Bahri (36°48′ 4.58″N, 3°34′ 7.01″E), Cap Djinet (36°52′ 37″ N, 3°43′ 23″E), Bouharoun (36′370 24″ N, 2°39′ 17″ E) and Cherchell (36°36′ 31″ N, 2°11′ 50″ E), Algeria (this paper).
Site on host: pharyngeal tooth pads, gills.
Prevalence and intensity: ex S. aurata off Algeria: 34% (31 out of 90 fish).
Specimens from Algeria, from S. aurata (Figures 1 and 4): vouchers deposited in the collections of the Swedish Museum of Natural History, Stockholm (SMNH-208363–SMNH-208373; SMNH-218760–SMNH-218780).
Specimens with molecular information: entire specimen lacking only a small lateral part (below the haptor) parts of specimens mounted on slide, lateral excised part used for molecular analysis: specimens from Algeria, from S. aurata: SMNH-208 363 (GenBank accession numbers OR149163, OR148271), SMNH-208 364 (GenBank accession numbers OR187608, OR148272), SMNH-208 365 (GenBank accession numbers OR148273).
Redescription
Measurements and description based on 33 stained and mounted specimens (Table 4). Body stocky, 1727 ± 267 (1106– 2280, n = 32) long, 693 ± 143 (447–1072, n = 32) maximum width at posterior third of body proper (Figure 4B). Two prohaptoral suckers muscular, 138 ± 33 (82–192) in diameter, oval, embedded in a lobed pad located in anterolateral margin of anterior region. Pharynx pyriform muscular. Two pairs of eyespots visible at level of pharynx. Intestinal ceca branched, with few lateral diverticula, mostly obscured by vitellarium, confluent posteriorly.
Table 4. Measurements of E. vallei from different hosts and localities.
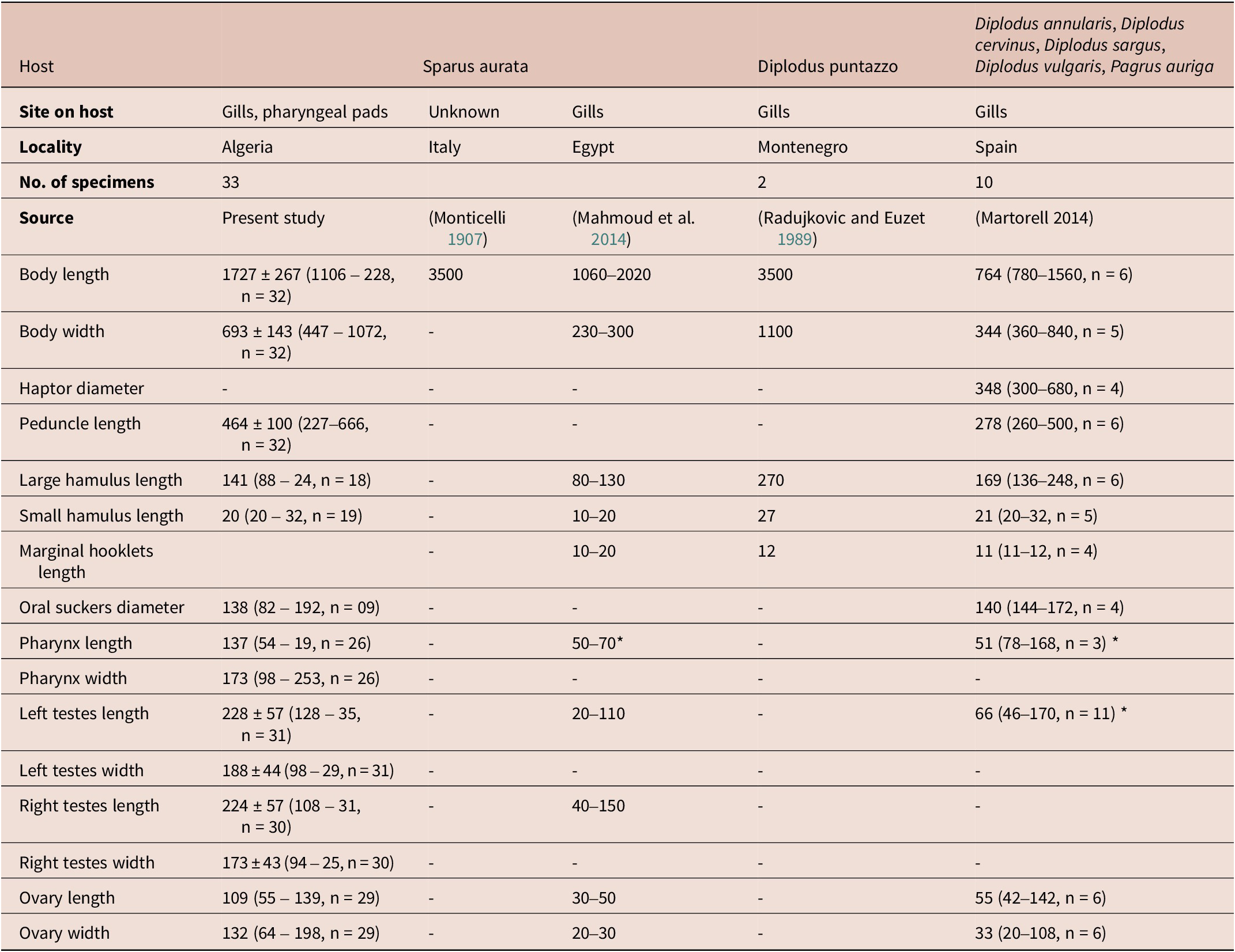
* Diameter.
Haptor bell-shaped, without septa; peduncle 464 ± 100 (227–666, n = 32) in length. Haptor armed with a pair of large hamuli (Figure 4C), a pair of smaller hamuli (Figure 4D) and 14 marginal hooklets. Length of median haptoral sclerites as follows: large hamulus, 141 ± 50 (88–246, n = 18) in length; small hamulus, 214 (84–289, n = 19) in length. Testes rounded, relatively large and unequal in size, located at midlevel of the body proper, length 228 ± 57 (128–356, n = 31); width 188 ± 44 (98–299, n = 31) for left testes and 224 ± 57 (108–319, n = 30) length, 173 ± 43 (94–258, n = 30) width for right testes (Figure 4). Vasa efferentia not observed. Vas deferens thick-walled, becoming convoluted alongside transverse vitelloducts, entering cirrus sac and enlarging into a fusiform internal seminal vesicle. Cirrus housed in cirrus sac, receiving posteriorly ejaculatory duct. Cirrus sac fusiform, muscular. Ovary anterior to tests, oval, giving rise to a long oviduct, length 109 (55–139, n = 29), 132 (64–198, n = 29) width. Ovo-vitelline duct and oötype not observed. Vitelline reservoir large. Vitellarium muscular, extending from level of ejaculatory duct to posterior end of body proper. Eggs not observed.
Discussion
To date, the genus Encotyllabe comprises 24 species, all of which are parasites of marine fish belonging to various families. They are found in a wide range of aquatic environments, including temperate, subtropical and tropical waters (Lebedev Reference Lebedev1967; WoRMS 2023). Among these species, E. vallei is common in sparid fish species as indicated in Table 5. First described from S. aurata, it was reported on sparids mainly of the genus Diplodus (Martorell Reference Martorell2004). There is also a single mention of its occurrence on the common dentex D. dentex (Linnaeus, 1758) (Euzet et al. Reference Euzet, Combes and Caro1993), Dentex sp. (Palombi, Reference Palombi1949) and on the redbanded seabream Pagrus auriga Valenciennes, 1843 (Martorell Reference Martorell2004).
Table 5. Hosts and localities of E. vallei Monticelli, Reference Monticelli1907. All localities are from the Mediterranean.

1 Reported as E. spari..
Furthermore, there have been records of E. vallei being on the striped red mullet, M. surmuletus Linnaeus, collected off the coast of Spain. However, the authors of this report suggested an accidental parasitism given that both the sparid type-host (S. aurata) and the mullet host (M. surmuletus) share the same habitat. Alternatively, it could potentially indicate the existence of a distinct species (Ferrer-Castello Reference Ferrer-Castello2015). Most interestingly, in the figure’s caption, Ferrer Castelló (2015) additionally referred to four individuals of Encotyllabe found on the annular sea bream Diplodus annularis (Linnaeus, 1758) but did not discuss it further. Curiously, in previous investigations of the transmission of monogeneans in farmed fish, the authors noted the absence of transmission of E. vallei from S. aurata to D. labrax (Fernandez-Jover et al. Reference Fernandez-Jover, Faliex, Sanchez-Jerez, Sasal and Bayle-Sempere2010; Papoutsoglou Reference Papoutsoglou2016).
Mahmoud et al. (Reference Mahmoud, Mahmoud and Fahmy2014) attributed the capsalid Monogenea collected on S. aurata off Egypt, which is the usual host and the type-host of E. vallei to E. spari (Mahmoud et al. Reference Mahmoud, Mahmoud and Fahmy2014). However, a closer examination and comparison of the photograph of their specimens with the illustration of E. spari provided by Yamaguti (Reference Yamaguti1934) reveals slight differences in the position of tests and shape of large hamuli (see Figure 1 in Mahmoud et al. (Reference Mahmoud, Mahmoud and Fahmy2014) versus Figure 7 provided by Yamaguti (Reference Yamaguti1934)). Additionally, E. spari was first described from three different hosts: the blackhead seabream Acanthopagrus schlegelii (Bleeker.) (jun. syn. Sparus macrocephalus (Basilewsky)), the trout sweetlips Plectorhinchus pictus (Tortonese) and Hong Kong grouper Epinephelus akaara (Temminck and Schlegel) from a distinct locality off Japan. It is highly likely that the capsalids reported by Mahmoud et al. (Reference Mahmoud, Mahmoud and Fahmy2014) are E. vallei.
We note that some authors referred to the ‘absence’ of a detailed redescription of E. vallei (Sepúlveda et al. Reference Sepúlveda, González and Oliva2014; Taborda et al. Reference Taborda, Sepulveda, Luque, Escribano and Oliva2023). However, it should be noted that Radujkovic and Euzet (Reference Radujkovic, Euzet, Radujkovic and Raibaut1989) did indeed provide a detailed and beautifully illustrated redescription of E. vallei, even though their specimens were collected from Montenegro and not from the type-host. Furthermore, it was sometimes mentioned that Monticelli reported E. vallei on S. aurata and Dentex sp. (Radujkovic and Euzet Reference Radujkovic, Euzet, Radujkovic and Raibaut1989; Zakia Reference Zakia2019). However, it is important to clarify that the original description of E. vallei was based on specimens collected from S. aurata, and the first mention on Dentex sp. was made by Palombi (Reference Palombi1949). The illustrations and morphometric data, along with the molecular data generated herein, will provide satisfactory and reliable material for comparison and for understanding the patterns of host specificity of this monogenean.
The first 28S rDNA and cox1 sequences for species of Encotyllabe in the Mediterranean and of E. vallei were provided by Lablack et al. (Reference Lablack, Rima, Georgieva, Marzoug and Kostadinova2022), who conducted an extensive and valuable data collection of monogeneans from the southwestern Mediterranean. However, some issues arise when analyzing all records of Encotyllabe from sparids from Algeria:
In their study, Lablack et al. (Reference Lablack, Rima, Georgieva, Marzoug and Kostadinova2022) proposed the existence of two putative species within the genus Encotyllabe based on genetic divergence in the cox1 gene. The first putative species, referred to as Encotyllabe sp. 1, was found on Pagellus bogaraveo, while the second putative species, Encotyllabe sp. 2, was found concurrently on D. vulgaris and S. aurata. They observed a range of genetic divergence within these species that fell between 0% and 1.9%.
Lablack et al. (Reference Lablack, Rima, Georgieva, Marzoug and Kostadinova2022) considered this divergence to be ‘slightly higher’ than the upper limit of the intraspecific ranges observed in other Encotyllabe species, specifically referring to E. antofagastensis and E. cheilodactyli.
In light of the available data and taking into consideration the molecular data generated so far, it can be deduced that Encotyllabe sp. 1 is oioxenic,Footnote 1 meaning it is highly specific and primarily associated with P. bogaraveo. On the other hand, Encotyllabe sp. 2 appears to be stenoxenic, indicating it exhibits a narrower host range, primarily infecting D. vulgaris and S. aurata.
A similar situation had been demonstrated for microcotylids from sparids: Microcotyle visa Bouguerche, Gey, Justine and Tazerouti, Reference Bouguerche, Tazerouti, Gey and Justine2019 and M. isyebi Bouguerche, Gey, Justine and Tazerouti, Reference Bouguerche, Tazerouti, Gey and Justine2019 are oioxenic to their hosts, Pagrus caeruleostictus (Valenciennes.) and Boops boops (Linnaeus.) (Bouguerche et al. Reference Bouguerche, Gey, Justine and Tazerouti2019a, Reference Bouguerche, Gey, Justine and Tazerouti2019b; Víllora-Montero et al. Reference Víllora-Montero, Pérez-del-Olmo, Georgieva, Raga and Montero2020); Microcotyle whittingtoni Víllora‑Montero, Pérez‑del‑Olmo, Georgieva, Raga and Montero, 2020 is oioxenic to D. dentex (Víllora-Montero et al. Reference Víllora-Montero, Pérez-del-Olmo, Georgieva, Raga and Montero2020), while the enigmatic M. erythrini Van Beneden et Hesse, 1863 is rather stenoxenic, occurring on Pagellus erythrinus (Linnaeus) and on P. pagrus (Linnaeus) (Víllora-Montero et al. Reference Víllora-Montero, Pérez-del-Olmo, Georgieva, Raga and Montero2020). Hence, we could assume that Encotyllabe sp. 1 is oioxenic while Encotyllabe sp. 2 is stenoxenic, but a more significant concern arises when trying to distinguish between Encotyllabe sp. 1 and Encotyllabe sp. 2. (1.1–1.9%) is well below the threshold for Monogenea and invertebrates, and higher divergences were used to separate Encotyllabe species (see Table 6). In the present analysis, the divergence of E. vallei from S. aurata ranged between 0–2 % which suggests that a single species is present in S. aurata. Similarly, the divergence between Encotyllabe from S. aurata, D. vulgaris and P. bogaraveo does not exceed 2%, below the interspecific divergence for cox1 known for Monogenea. We thus suggest that E. vallei is the sole species infesting the previously mentioned host.
Table 6. Intraspecific and interspecific variations of the cox1 gene within species of Encotyllabe

It is important to note that stenoxenic specificity is not unusual in Monogenea and has been previously demonstrated using molecular barcoding (Bouguerche et al. Reference Bouguerche, Gey, Justine and Tazerouti2019a, Lablack et al. Reference Lablack, Rima, Georgieva, Marzoug and Kostadinova2022; Víllora-Montero et al. Reference Víllora-Montero, Pérez-del-Olmo, Georgieva, Raga and Montero2020). For instance, the microcotylid M. erythrini was described from the sparid Pagellus erythrinus off Brest (Brittany, Atlantic Ocean) than recorded from three other hosts (Pagellus acarne, B. boops and D. dentex) in several localities in the Mediterranean (Bouguerche et al. Reference Bouguerche, Gey, Justine and Tazerouti2019b). Despite the record of M. erythrini sensu lato from the record from B. boops being demonstrated to be a distinct species, M. erythrini sensu stricto had been shown to be exceptionally a stenoxenic microcotylid, occurring on P. erythrinus and P. pagrus (demonstrated by cox1 barcoding) (Víllora-Montero et al. Reference Víllora-Montero, Pérez-del-Olmo, Georgieva, Raga and Montero2020). Encotyllabe vallei presents another ‘enigmatic’ monogenean species, displaying a host specificity pattern distinct from that of its congeners.
In our current study, it is noteworthy that the molecular tree based on 28S sequences illustrates the monophyly of three subfamilies of capsalids: the Encotyllabinae, Benedeniinae and Capsalinae (Figure 2). Perkins et al. (2009), in a molecular study based on different markers, retrieved only the Capsalinae as monophyletic. Similarly, Gastineau et al. (Reference Gastineau, Bouguerche and Tazerouti2023) retrieved a monophyly of the Capsalinae, with representatives of all four genera of the subfamily (Capsala Bosc, 1811; Capsaloides Price, 1936; Tristoma Cuvier, 1817 and Nasicola Yamaguti, 1968) united together in a well-supported monophyletic clade. Results for other subfamilies were less in accordance with traditional taxonomy (Gastineau et al. Reference Gastineau, Bouguerche and Tazerouti2023). Hence, future phylogenetic investigations, using more extensive datasets, will certainly offer a more comprehensive insight into the evolutionary history of capsalid monogeneans.
The distribution of Encotyllabe spp. according to the systematics of host groups is given in Table 1, which summarizes updated data originally compiled by Lebedev (Reference Lebedev1967). From the available data, a discernible pattern emerges in the geographical distribution of Encotyllabe species: Encotyllabe lintoni Monticelli, 1909; Encotyllabe monticelli Perez Vigueras, 1940 and Encotyllabe pricei Koratha, 1955 occur in North America. E. pagrosomi MacCallum, Reference MacCallum1917 and Encotyllabe embiotocae Noble, Reference Noble1966 occur in Pacific waters; E. chironemi is restricted to the coast of New Zealand and in the seas of Japan; Encotyllabe caranxi Lebedev, Reference Lebedev1967 described from New Zealand-Australian fish; E. pagelli is known from the coast of Belgium and Ireland; E. spari Yamaguti, Reference Yamaguti1934; E. lutiani Tripathi, Reference Tripathi1959 and E. masu Ishii and Sawada, Reference Ishii and Sawada1938 is found off the coast of India. The Mediterranean species are E. vallei, E. paronae and E. nordmanni. The validity of all the Mediterranean species in addition to E. pagelli was questioned (Taborda et al. Reference Taborda, Sepulveda, Luque, Escribano and Oliva2023). Another striking aspect when analyzing these data is that nearly all Encotyllabe spp. were described from a single host species or closely related host species, as previously noted by Taborda et al. (Reference Taborda, Sepulveda, Luque, Escribano and Oliva2023), suggesting host specificity. However, the findings presented in our study challenge the notion of strict host specificity for Encotyllabe, particularly for E. vallei indicating that it primarily associates with a narrower but still specific range of host species. A similar conclusion was reached for Encotyllabe species from Haemulidae hosts, as such Encotyllabe haemuli and E. antofagastensis which formed a well-supported clade (see Taborda et al. (Reference Taborda, Sepulveda, Luque, Escribano and Oliva2023)).
Determining the taxonomic status of Encotyllabe spp. presents a challenging task, as traditional taxonomy relies on a variety of morphological characteristics, including body shape, the relative sizes of several organs, the shape and relative position of the testes, the penis shape, the extension of the vitellaria and the size and shape of the anchors, as well as the relative distances between different organs (Robinson Reference Robinson1961; Sepúlveda et al. Reference Sepúlveda, González and Oliva2014; Taborda et al. Reference Taborda, Sepulveda, Luque, Escribano and Oliva2023). However, it is important to note that the presence of an evaginated or protrusible pharynx, as suggested by Martorell (Reference Martorell2004), adds a layer of complexity to this taxonomic classification. Considering this, it may be advisable to exclude the size of the pharynx as a definitive criterion for taxonomic differentiation.
Because P. bogaraveo is the type-host of E. pagelli (Van Beneden and Hesse Reference Van Beneden and Hesse1863), we were at first tempted to suggest a synonymy between E. vallei and E. pagelli. However, we refrained from doing so as E. pagelli was first described from the Atlantic coast off Brest, France. In the absence of available morphological data for Encotyllabe from P. bogaraveo, it remains challenging to definitively determine the host specificity of E. vallei. There is the possibility that a species complex exists within individual sparid fish species, as has been observed in some other Encotyllabe species. The acquisition of morphometric and morphological data from specimens collected from P. bogaraveo would be instrumental in shedding light on the host specificity patterns of this capsalid and clarifying its taxonomic status within the genus.
Conflict of interest declaration
The authors declare that they have no competing interests.
Author contributions
Fatima-Zohra Zedam: conceptualisation; data curation; investigation; methodology; project administration; validation; writing – original draft. Chahinez Bouguerche: data curation; funding acquisition; investigation; methodology; project administration; validation; writing – original draft, review and editing. Mohammed Ahamed: conceptualisation; data curation; investigation; methodology; project administration; validation. Fadila Tazerouti: project administration. All authors read and approved the final manuscript.
Acknowledgements
This study is wholeheartedly dedicated to Professor Nadia Kechemir-Issad, an Algerian parasitologist and a former student of the late Claude Combes, a French biologist and parasitologist for her passionate teaching of host-parasite interactions. We thank members of the Department of Zoology from the Swedish Museum of Natural History, Sweden, especially Dr. Oleksandr Holovachov for kindly providing accession numbers. Our thanks are due to fishermen from Algiers. We thank Professor Jean-Lou Justine from Institut de Systématique, Évolution et Biodiversité (ISYEB), Muséum National d’Histoire Naturelle (MNHN), Paris, France, for providing comments on the manuscript. This research was supported by the Department of Zoology, Swedish Museum of Natural History, Stockholm, Sweden.
Financial support
Chahinez Bouguerche was supported individually by the Swedish Taxonomy Initiative, Artdatabanken, Swedish University of Agricultural Sciences within the scope of the project ‘Taxonomy and systematics of digenetic trematodes parasitizing fishes of Sweden’ (dha 2019.4.3-48). The funders had no role in study design, data collection and analysis, the decision to publish or the preparation of the manuscript.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation. The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.












