Introduction
Fascioliasis is an important zoonotic parasite infection caused by digenean trematodes of the genus Fasciola, and the common species are Fasciola hepatica Linnaeus (1758) and F. gigantica Cobbold (1856) (Mas-Coma et al., Reference Mas-Coma, Valero and Bargues2009, Reference Mas-Coma, Bargues and Valero2018; Mochankana and Robertson, Reference Mochankana and Robertson2016). The disease is distributed worldwide (Hurtrez-Boussès et al., Reference Hurtrez-Boussès2001; Mas-Coma, Reference Mas-Coma2005), affecting mainly domestic and wild ruminants, but also humans (Walker et al., Reference Walker2008; Robinson and Dalton, Reference Robinson and Dalton2009; Phalee et al., Reference Phalee2015; Mas-Coma et al., Reference Mas-Coma, Bargues and Valero2018), causing economic loss in livestock production globally (Khan et al., Reference Khan2013). Of the two Fasciola spp., F. hepatica has a cosmopolitan distribution, with human infection reported in five continents (Hurtrez-Boussès et al., Reference Hurtrez-Boussès2001; Mas-Coma, Reference Mas-Coma2005; Mas-Coma et al., Reference Mas-Coma, Valero and Bargues2009, Reference Mas-Coma, Bargues and Valero2018), whereas F. gigantica is more limited to Africa and Asia (Mas-Coma et al., Reference Mas-Coma, Valero and Bargues2009, Reference Mas-Coma, Bargues and Valero2014, Reference Mas-Coma, Bargues and Valero2018; Abebe et al., Reference Abebe2010), occurring to a lesser extent in southern parts of Europe, Turkey, the Near East and some southern states of the old USSR, Armenia in particular (Mas-Coma, Reference Mas-Coma2005), and South America (Hurtrez-Boussès et al., Reference Hurtrez-Boussès2001). Furthermore, distribution of both species also overlaps in many areas in Africa and Asia (Mas-Coma et al., Reference Mas-Coma, Bargues and Valero2005).
Fasciola spp. require freshwater snails as the intermediate host (IH) to complete their life cycle, and mammals, mainly herbivores, including humans, act as the definitive hosts (Mas-Coma et al., Reference Mas-Coma, Bargues and Valero2005). Freshwater snails of the family Lymnaeidae act as the IHs of F. hepatica and F. gigantica (Bargues and Mas-Coma, Reference Bargues and Mas-Coma2005; Caron et al., Reference Caron2014), and play a significant role in determining the distribution and epidemiology of fascioliasis (Mochankana and Robertson, Reference Mochankana and Robertson2016). Hence, the distribution pattern and abundance of the two species is equally associated with and influenced by the global distribution of their lymnaeid intermediate snail host (Mas-Coma et al., Reference Mas-Coma, Bargues and Valero2005). Fasciola gigantica utilizes Radix species (Mas-Coma, Reference Mas-Coma2005), particularly species belonging to the Lymnaea (R.) auricularia (Linnaeus, 1758) complex (Hubendick, 1951) as IHs (Brown, Reference Brown1994), whilst Lymnaea (Radix) natalensis Krauss (1848) is considered to be the main IH of F. gigantica in Africa (Walker et al., Reference Walker2008). In contrast, F. hepatica uses snail species from the genus Galba/Fossaria as IHs (Mas-Coma, Reference Mas-Coma2005). However, the geographical distribution of F. hepatica is mainly linked to the geographical expansion of its original European lymnaeid IH Lymnaea (Galba) truncatula (Müller, 1774), the spread of the American species Lymnaea (Pseudosuccinea) columella (Say, 1817), and its adaptation to other lymnaeid species native to the newly colonized areas (Mas-Coma et al., Reference Mas-Coma, Bargues and Valero2005). In South Africa, the presence of G. (L.) truncatula has been reported and has been assumed to be transmitting F. hepatica in areas where it has been found (De Kock et al., Reference De Kock, Wolmarans and Bornmas2003).
According to Quayle et al. (Reference Quayle, Appleton and Dickens2010) and Mucheka et al. (Reference Mucheka2015), both F. hepatica and F. gigantica are present in South Africa, and L. (G.) truncatula and L. (R.) natalensis act as their IHs, respectively. Despite the overlap in the distribution of both Fasciola species observed by Mucheka et al. (Reference Mucheka2015) in KwaZulu-Natal and Mpumalanga provinces, both F. hepatica and F. gigantica seem to have different altitudinal distribution in accordance to the ecological requirements of their respective snail IHs in the country (Quayle et al., Reference Quayle, Appleton and Dickens2010). Galba (L.) truncatula is found in cooler areas and is mostly abundant in high areas such as Lesotho and the northern parts of the Eastern Cape province of South Africa (De Kock et al., Reference De Kock, Wolmarans and Bornmas2003; Quayle et al., Reference Quayle, Appleton and Dickens2010), whereas L. (R.) natalensis is one of the most widely distributed snail species according to the National Freshwater Snail collection (NFSC) records (De Kock et al., Reference De Kock, Joubert and Pretorius1989) but is not found in the cooler, drier regions of South Africa (Brown, Reference Brown1994; Quayle et al., Reference Quayle, Appleton and Dickens2010).
Lymnaea (P.) columella is a rapidly colonizing, heat tolerant species thought to have originated from Central America, the Caribbean, and the southern part of North America (Mas-Coma et al., Reference Mas-Coma, Bargues and Valero2005). The species is considered to be invasive in southern Africa (Appleton, Reference Appleton2003), including South Africa, where it has been considered one of the most successful colonists of freshwater species in the country (De Kock et al., Reference De Kock, Joubert and Pretorius1989), and one of the most widespread freshwater snails by the NFSC, following L. (R.) natalensis and Bulinus tropicus (De Kock et al., Reference De Kock, Joubert and Pretorius1989). Lymnaea (P.) columella has been documented to act as an intermediate host of both F. hepatica and F. gigantica in many countries (Mas-Coma et al., Reference Mas-Coma, Bargues and Valero2005; Grabner et al., Reference Grabner2014). Although this invasive species has been presumed to be susceptible to both flukes in South Africa (Brown, Reference Brown1980), the epidemiological role played by this species in the transmission of the two Fasciola species has not been proven. Hence, this study screened L. (P.) columella snails collected from selected locations of KwaZulu-Natal and Eastern Cape provinces of South Africa known to be endemic for fascioliasis in cattle and sheep to determine the infection status with F. hepatica and/or F. gigantica using molecular techniques.
Materials and methods
Snail collection and identification
Lymnaea (Pseudosuccinea) columella snails were collected in November 2017 and July 2018 from two locations at Hazelmere dam (HD1 and HD2) in Verulam area in KwaZulu-Natal province, at elevations ranging from 91 to 92 m above sea level, and from three locations at Lucingweni (LC1, LC2 and LC3) in Mthatha area in Eastern Cape province, at elevations ranging from 717 to 800 m above sea level, using a metal scoop (30 × 30 cm) (fig. 1). Snails were identified as L. (P.) columella following descriptions by Brown (Reference Brown1994). Immediately after collection, more than 50 snails were screened for cercariae shedding as described by Sharif et al. (Reference Sharif, Daryani and Karimi2010). After screening, a total of 20 snails per location (n = 100) were selected randomly and fixed in 70% ethanol for molecular analysis.
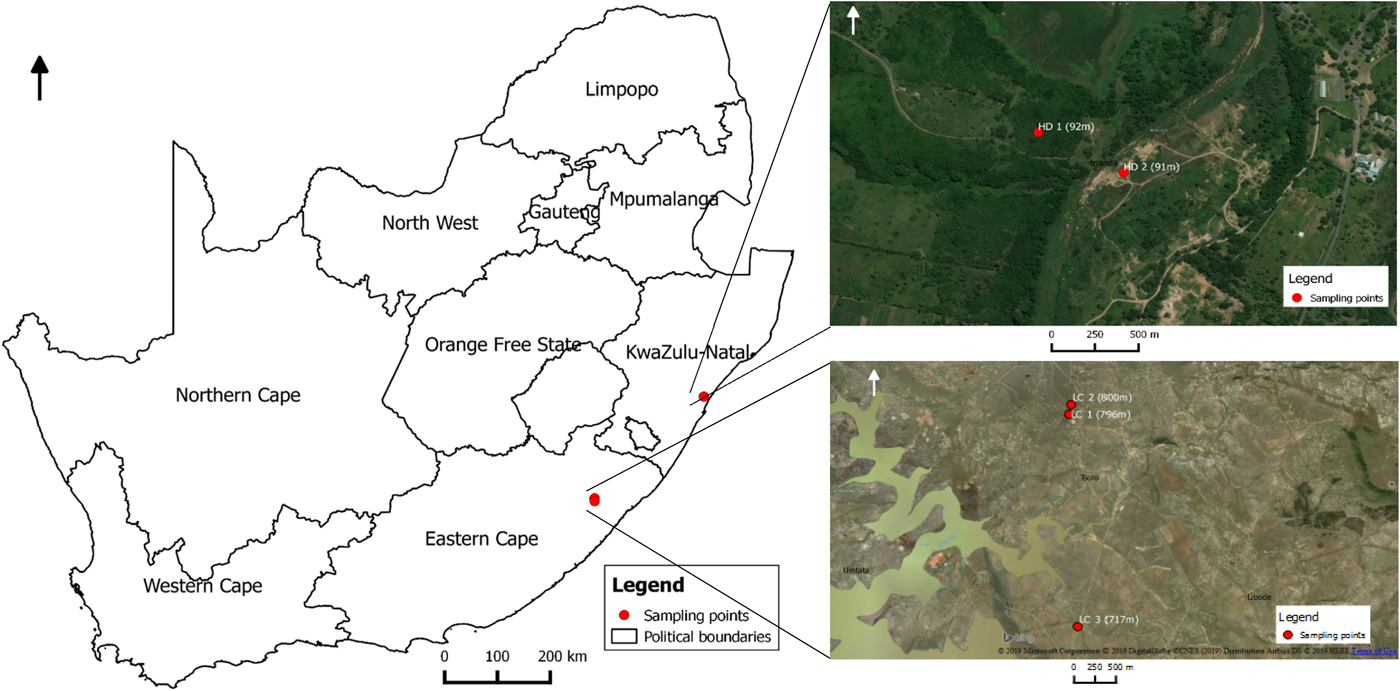
Fig. 1. Map showing locations were Lymnaea (Pseudosuccinea) columella were collected from KwaZulu-Natal and Eastern Cape provinces of South Africa.
Molecular identification of Fasciola spp. in L. (P.) columella tissue
DNA was extracted from the homogenized individual L. (P.) columella snail tissue using a Genomic DNA™ Tissue MiniPrep Kit (Zymo Research Corporation, Irvine, CA, USA) according to the manufacturer's instructions. In order to identify infected snails, and distinguish Fasciola species within the snails, the ITS-1 region of Fasciola spp. was amplified using the primers FascF: 5′-ACCGGTGCTGAGAAGACG-3′ and FascR: 5′-CGACGTACGTGCGTCCA-3′ (Rokni et al., Reference Rokni2010). Polymerase chain reaction (PCR) amplification was performed in a 25 μl reaction volume, each containing 9.5 μl sterile water, 12.5 μl PCR Master Mix (2X) (Thermo Fisher Scientific, Waltham, MA, USA), 0.5 μl (10 μm) of each primer and 2 μl of genomic DNA. Amplification was performed under the following thermocycling conditions: 95°C for 5 minutes as initial denaturation, followed by 40 cycles of (denaturation at 95°C for 45 s, annealing at 60°C for 45 s and extension at 72°C for 1 minute), and lastly 72°C for 7 minutes as final extension. Fragments were separated by 2% agarose gel electrophoresis stained with ethidium bromide, then visualized and photographed in a transilluminator (BIORAD).
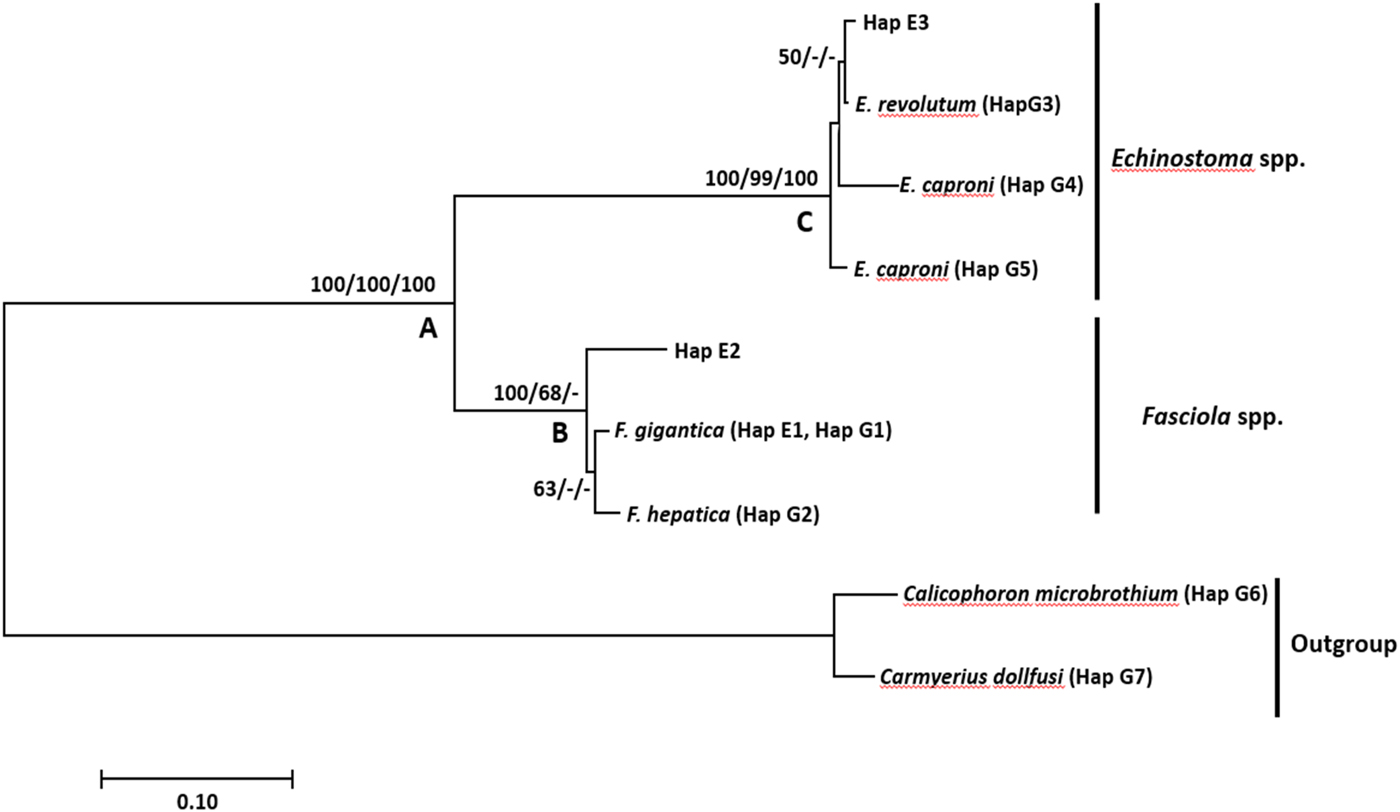
Fig. 2. Neighbour-joining tree based on the nuclear ribosomal ITS-1 region illustrating the relationship between Fasciola gigantica, F. hepatica and Echinostoma spp. from Lymnaea (Pseudosuccinea) columella (Hap E 1–3) collected from Eastern Cape and KwaZulu-Natal provinces of South Africa, and the close matches from the NCBI GenBank and outgroups (G 1–7). Nodal support values are shown in the order of neighbour-joining bootstrap value, Bayesian inference probability and maximum likelihood bootstrap value.
Sequencing and sequence analysis
To confirm the identity of Fasciola spp., amplicons from positive snails were sent for sequencing at Inqaba Biotechnical Industries (Pty) Ltd. (Pretoria, South Africa). The obtained sequences were assembled, edited and aligned with rDNA homologue sequences obtained from the GenBank database with Clustal W (Thompson et al., Reference Thompson1997) using the BioEdit program (Hall, Reference Hall1999). DnaSP (v 5.10.1) (Rozas et al., Reference Rozas2003) was used to generate and determine the number of haplotypes generated by the dataset. The most appropriate model test of nucleotide substitution for neighbour-joining (NJ), maximum likelihood (ML) and Bayesian inference analyses were selected using jModeltest (Posada, Reference Posada2008). The SYM model was selected under the AIC information criterion. Neighbour-joining and maximum likelihood trees were generated using PAUP* 4.0 (Swofford, Reference Swofford2002). The nodal support for both methods was estimated using 1000 bootstrap pseudo-replicates. Bayesian inference was carried out in MrBayes 3.1.2 (Huelsenbeck and Ronquist, Reference Huelsenbeck and Ronquist2001). Four Markov chains were run for 5 million generations to ensure that the standard deviation of the split frequencies was < 0.01. The first 500,000 trees were discarded as burn in, after an initial run to check that this was sufficient to achieve stationarity.
Results
All 100 snails from the five locations screened for infection by shedding of cercariae were negative. When PCR was conducted based on an ITS-1 marker designed specifically for identification of Fasciola spp., all the 100 snails (20 per site) were positive (table 1), and positivity was denoted by a band of approximately 460 base pairs. Blast analysis of five sequenced samples per location (n = 25) showed that sequences from snails collected from the LC1, LC2 and LC3 locations in Mthatha area gave a 99% homology with a complete appropriate region of the ribosomal nuclear sequence of F. gigantica (accession number MG201868.1) (table 1). Furthermore, four snails from the HD1 and HD2 locations in Verulam area harboured F. gigantica and one snail in each location had Echinostoma spp., with 93–99% homology for F. gigantica (accession number MG201868.1) and 99% homology with Echinostoma revolutum (table 1).
Table 1. Percentage similarities of Fasciola and Echistostoma species obtained intra Lymnaea (Pseudosuccinea) columella from selected locations in KwaZulu-Natal and Eastern Cape provinces, and their closest GenBank-derived isolates.

HD 1, Hazelmere dam location 1; HD 2, Hazelmere dam location 2; LC 1, Lucingweni location 1; LC 2, Lucingweni location 2; LC 3, Lucingweni location 3
The F. gigantica sequences from this study were 420 nucleotide bases long and homogenous except for one Fasciola species from HD2 in Verulam area, KwaZulu-Natal province. The haplotype analysis yielded nine haplotypes consisting of the samples derived from GenBank and isolates from our study (table 2). Haplotype one (Hap E1) was the most common, comprising 22 isolates from this study and six F. gigantica isolates from GenBank (table 2). Phylogenetic analysis showed that the outgroup formed a strongly supported monophyletic clade (clade A) with the ingroup samples (fig. 1). Clade A further showed a distinction between the Fasciola species (clade B) and Echinostoma species (clade C). Clade B, which was strongly supported by neighbour-joining and moderately supported by maximum likelihood, included all Fasciola species. Within this clade (B), there was a distinction between F. gigantica and F. hepatica isolates, which formed a moderately supported monophyletic clade by neighbour-joining, with isolate Hap E2 (KZ2.1) from HD2 location in Verulam area branching out forming a sister clade to the two Fasciola species. A strongly supported clade C consisted of two isolates (Hap E6), each from HD1 and HD2 locations in Verulam area and three Echinostoma isolates from GenBank, namely E. caproni (KF425322.1, U58098.1) and E. revolutum (U58102.1).
Table 2. Haplotype status of Fasciola spp. and Echinostoma spp. from Verulam area in KwaZulu-Natal, and Mthatha area in Eastern Cape, and the GenBank-derived isolates.

E, experimental samples; G, GenBank-derived isolates; E, Eastern Cape province; KZ, KwaZulu-Natal province
Discussion
The current study demonstrated a high rate of natural infection of L. (P.) columella with F. gigantica from Mthatha area in Eastern Cape and Verulam area in KwaZulu-Natal provinces in South Africa. Based on reports elsewhere this invasive lymnaeid species acts as an intermediate host of both F. gigantica and F. hepatica (Bargues and Mas-Comas, Reference Bargues and Mas-Coma2005). Confirmation of presence of this snail species in South Africa led to the assumption that the observed increase in the prevalence of fascioliasis was due to the introduction of L. (P.) columella (De Kock et al., Reference De Kock, Joubert and Pretorius1989), and no information is available confirming natural infection of L. (P.) columella with F. gigantica. With both Fasciola species occurring in South Africa and F. hepatica being reported as the most prevalent species in KwaZulu-Natal by Mucheka et al. (Reference Mucheka2015), it is surprising that none of the P. columella snails collected from the two locations were infected with F. hepatica.
Lymnaea (Pseudosuccinea) columella is widely distributed in South Africa, ranking as the third most distributed freshwater snail in the country, surpassed only by L. (R.) natalensis and Bulinus tropicus (De Kock et al., Reference De Kock, Joubert and Pretorius1989). Furthermore, this species has been found to occur frequently in the same habitats as L. (R.) natalensis (De Kock et al., Reference De Kock, Joubert and Pretorius1989; Wolmarans and De Kock, Reference Wolmarans and De Kock2006; Kemp et al., Reference Kemp2016) and B. tropicus (De Kock et al., Reference De Kock, Joubert and Pretorius1989). The ability of L. (P.) columella to adapt and tolerate various ecological and climatic conditions and to coexist with L. (R.) natalensis (main IH of F. gigantica) allows the snail to transmit the two Fasciola species (De Kock et al., Reference De Kock, Joubert and Pretorius1989). The invasive nature of this lymnaeid species, and its ability to invade in areas with summer and winter rainfall, cold highveld regions such as the Transvaal area and Free State, and the hot subtropical regions of the Transvaal Lowveld and KwaZulu-Natal (De Kock et al., Reference De Kock, Joubert and Pretorius1989), gives this species advantage over the natural intermediate hosts of both F. hepatica (G. (L.) truncatula) and F. gigantica (L. (R.) natalensis). Hence, the high rate of infection of L. (P.) columella with F. gigantica as shown in this study is an indication of the significant role played by this snail species in extending the geographical distribution of the parasite in South Africa.
Results showed that isolates of F. gigantica from our study formed one haplotype with the GenBank-derived isolates. The phylogenetic tree showed that F. hepatica formed a moderately supported sister clade to F. gigantica. This is similar to the findings of Mucheka et al. (Reference Mucheka2015), although these were strongly supported. The sequence analysis supported the identification of 22 isolates—15 from Mthatha and seven from Verulam (4 from HD1 and 3 from HD2)—as F. gigantica, and one isolate (Hap E7) from HD2 as Fasciola sp., which formed a separate subclade from F. gigantica and F. hepatica. Furthermore, the isolate is separated from F. gigantica and F. hepatica by a mean genetic distance of 4.61% and 6.58%, respectively. Although this isolate seemed to be closely related to F. gigantica based on BLAST analysis with genetic p-distance, eight mutations were found between the isolate and F. gigantica in positions 79, 133, 195, 196, 197, 368, 376 and 377 of the alignment. The formation of a separate subclade of this isolate from the two Fasciola species leads to the suggestion of the possibility of a hybrid between the two species.
Besides F. gigantica, Echinostoma sp. was also detected in two L. (P.) columella snails from HD1 and HD2 locations in Verulam, KZN. These isolates formed a strongly supported clade with E. caproni (KF425322.1, U58098.1) and E. revolutum (U58102.1). Within this clade, this isolate formed a weakly supported subclade with E. revolutum. Genetic distance between the two Echinostoma isolates from our study (Hap E3) and the GenBank isolates further confirmed the close relatedness between our isolates (Hap E3) and the GenBank isolates, which showed a genetic distance of 0.658% and 30.0% between our isolates (Hap3) and E. revolutum and E. caproni, respectively. The detection of Echinostoma sp. correlates with the report by Grabner et al. (Reference Grabner2014), who detected natural infection of F. gigantica, Echinostoma caproni and unidentified Echinostoma sp. in L. (P.) columella in Egypt using PCR. Lymnaeids act as the first IH of E. revolutum, whereas E. caproni utilize Biomphalaria and Bulinus spp. as first intermediate hosts (Fried and Huffman, Reference Fried and Huffman1996), and most likely use L. (P.) columella as the second intermediate host (Fried and Huffman, Reference Fried and Huffman1996; Grabner et al., Reference Grabner2014). Nonetheless, infection of L. (P.) columella with Echinostoma spp. has not been reported in South Africa and, to the best of our knowledge, this is the first report in the country.
In conclusion, our study confirmed that L. (P.) columella acts as an intermediate host of F. gigantica and Echinostoma sp. in South Africa. This is the first study reporting natural infection of L. (P.) columella with F. gigantica and Echinostoma sp. in South Africa. Although the susceptibility of L. (P.) columella to F. gigantica has been reported elsewhere, this is the first report confirming natural infection in South Africa. Given the invasive nature of L. (P.) columella (Appleton, Reference Appleton2003), further studies are needed to determine the role of this snail in extending the geographical distribution of F. gigantica and F. hepatica in South Africa. Furthermore, there is a need for a countrywide survey of this invasive species to assess the geographical distribution and infection status with F. gigantica and F. hepatica in various geographical regions of South Africa and also carry out laboratory-based experiments to compare the IH competence between L. (P.) columella and L. (R.) natalensis in areas where the two species coexist.
Author ORCIDs
M.P. Malatji, 0000-0002-6188-4204
Acknowledgements
The authors acknowledge students from the University of KwaZulu-Natal Parasitology Laboratory for assisting with snail collection.
Financial support
This study was supported financially by the National Research Foundation (NRF) of South Africa.
Conflict of interest
None.
Ethical standards
Experimental protocols for this study were reviewed and approved by the Animal Ethics Committee of the University of KwaZulu-Natal (Ref: AREC/044/016D) in accordance with the South African national guidelines on animal care, handling and use for biomedical research.






