Introduction
Cystic echinococcosis (CE) is a neglected disease caused by metacestode (larval) form of the Echinococcus granulosus sensu lato (sl) in humans. CE is mostly endemic in rural areas of Australia, Asia, South America and Mediterranean countries (Deplazes et al., Reference Deplazes, Rinaldi and Alvarez Rojas2017). Since it is mostly asymptomatic, definite prevalence of the disease is still unknown. However, the incidence of human CE is reported based on hospital records (Altintas, Reference Altintas2008). As reported in a recent study, CE prevalence is estimated to be approximately 0.61% in Turkey (Tamarozzi et al., Reference Tamarozzi, Akhan and Cretu2018). Although CE is mostly associated with a wide spectrum of symptoms, there is no specific finding. The hydatid cysts are mostly located in the liver and lung, with rates of about 70% and 25%, respectively (Akhan et al., Reference Akhan, Ozmen, Dinçer, Sayek and Göçmen1996; Brunetti et al., Reference Brunetti, Tamarozzi and Macpherson2018). Imaging techniques such as ultrasonography (US), computed tomography (CT) and magnetic resonance imaging (MRI) are extensively used for diagnosis of CE. Serological tests have complementary role to imaging modalities in diagnosis. The World Health Organization-Informal Working Group on Echinococcosis (WHO-IWGE) has published a widely accepted classification of liver hydatid cyst based on the activity of the disease. Based on this classification, hydatid cysts are divided into three clinical groups as active (CE1 and CE2), transitional (CE3a/CE3b) and inactive (CE4 and CE5) (Brunetti et al., Reference Brunetti, Kern and Vuitton2010; Kern et al., Reference Kern, Menezes da Silva, Akhan, Müllhaupt, Vizcaychipi, Budke and Vuitton2017). Although imaging techniques generally provide sufficient data for the diagnosis of CE, serological tests are needed in some cases. Unfortunately, there are considerable drawbacks in sensitivity/specificity (Se/Sp) and prognostic value of serological tests (Akhan & Ozmen, Reference Akhan and Ozmen1999; Manzano-Román et al., Reference Manzano-Román, Sánchez-Ovejero, Hernández-González, Casulli and Siles-Lucas2015). While several studies are reported that cyst characteristics affected CE serology, there is still limited data available on underlying mechanisms of the host during CE development and progression.
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules, that have function in RNA silencing and post-transcriptional regulation of gene expression. Some miRNA families are predominantly expressed in certain tissues while some others are specific to certain biological processes (Negrini et al., Reference Negrini, Nicoloso and Calin2009; Li et al., Reference Li, Li, Ding, He and Cheng2010). Unique circulating miRNA expression profiles have been demonstrated for various types of diseases. Mammalian miRNAs are known to be stable in extracellular fluids such as plasma, serum, urine, saliva and semen (Mitchell et al., Reference Mitchell, Parkin and Kroh2008; Olivieri et al., Reference Olivieri, Capri, Bonafè, Morsiani, Jung, Spazzafumo, Viña and Suh2017). With the discovery of the disease-specific miRNAs in the blood of patients with cancer, metabolic disorders or viral infections, miRNA expression profiles have been widely studied particularly in infectious diseases that are difficult to diagnose and follow-up. (Tritten et al., Reference Tritten, Burkman, Moorhead, Satti, Geary, Mackenzie and Geary2014). Besides, miRNAs have a crucial role in the regulation of host–pathogen relation due to their function as post-transcriptional mechanism regulators (Cai et al., Reference Cai, Gobert and McManus2016).
The aims of this study are (1) to determine the alterations in miRNA expression profiles of patients with active and inactive cysts compared with healthy controls and (2) to identify altered cellular pathways in CE patients.
Materials and methods
Ethics statement
This study was approved by the Institutional Ethical Committee of the Faculty of Medicine (GO 17/711-16).
Sample collection, RNA extraction and cDNA synthesis
Twenty confirmed CE patients with 23 CE cyst (13 female and seven male) and three healthy controls (one female and two male, mean age ±40, without any underlying chronic and infectious disease) were included in this study. During US examination, the WHO-IWGE classification was used to evaluate the cases (Brunetti et al., Reference Brunetti, Kern and Vuitton2010; Kern et al., Reference Kern, Menezes da Silva, Akhan, Müllhaupt, Vizcaychipi, Budke and Vuitton2017). Total RNA extraction was performed from serum of the patients/controls by miRNeasy Mini Kit (Qiagen) following the manufacturer's instructions. Quality and purity of the RNA was verified by a NanoDrop 2000c instrument (ThermoScientific). For cDNA synthesis, miScript II RT system (Qiagen) were used according to manufacturer's recommendations.
miRNA expression profiles and pathway analyses
miScript SYBR Green PCR Kit and miScript miRNA HC PCR Arrays (with a LightCycler 480 instrument II (Roche, Germany)) were used for the detection and quantification of miRNAs in serum. The miScript miRNA HC PCR Array provides expression profiles of 372 pathway/disease/functionally related mature miRNAs. Data analysis was performed using an online Geneglobe data analysis centre (https://geneglobe.qiagen.com/no/analyze/), which uses the comparative CT (ΔΔCT) method for relative quantification and indicates fold change calculations. p < 0.05 was considered as statistically significant.
For miRNA target prediction miRDB (http://mirdb.org/), Targetscan (http://www.targetscan.org/vert_72/), and DIANA Tools (http://diana.imis.athena-innovation.gr/DianaTools/index.php) were used (Friedman et al., Reference Friedman, Farh, Burge and Bartel2009; Paraskevopoulou et al., Reference Paraskevopoulou, Georgakilas, Kostoulas, Vlachos, Vergoulis, Reczko, Filippidis, Dalamagas and Hatzigeorgiou2013; Liu & Wang, Reference Liu and Wang2019; Chen & Wang, Reference Chen and Wang2020).
Results
Demographics of patients and clinical characteristics
The majority of the cases were female (65%, 13/20). Mean age of the patients were 37.7 (range 7–69 years).
Five of the 23 hydatid cysts were identified as CE1, four as CE2, one as CE3a, three as CE3b, seven as CE4 and three as CE5. Among 20 patients, 17 had a single cyst, the rest harboured two hydatid cysts. None of the patients had more than one cyst type. The mean size and volume of cysts were recorded as 7.4 cm and 251 cm3, respectively. All cysts were located in the liver (14/23 right lobe, 9/23 left lobe) (table 1). All the serum samples were positive for hydatidosis either by enzyme-linked immunosorbent assay (Hydatidosis IgG ELISA, Vircell SL, Granada, Spain) or indirect hemagglutination assay (Hydatidose, FUMOUZE Laboratories, France).
Table 1. Characteristics of patients.

miRNA expression profiles
In patients with active cysts, a total of 6 miRNA (hsa-miR-4659a-5p, hsa-miR-4518, hsa-miR-3977, hsa-miR-4692, hsa-miR-181b-3p, hsa-miR-4491) and in inactive CE patients only one miRNA (hsa-miR-4687-5p) expression was found to be downregulated by at least twofold (p < 0.05) compared with healthy controls. Relative expression profiles of these miRNAs in active and inactive patients are presented in figs 1 and 2.

Fig. 1. Heatmap of miRNA expressions among CE patients and healthy control groups. The map is produced by using the online GeneGlobe Data Analysis Center (https://geneglobe.qiagen.com/no/analyze/).
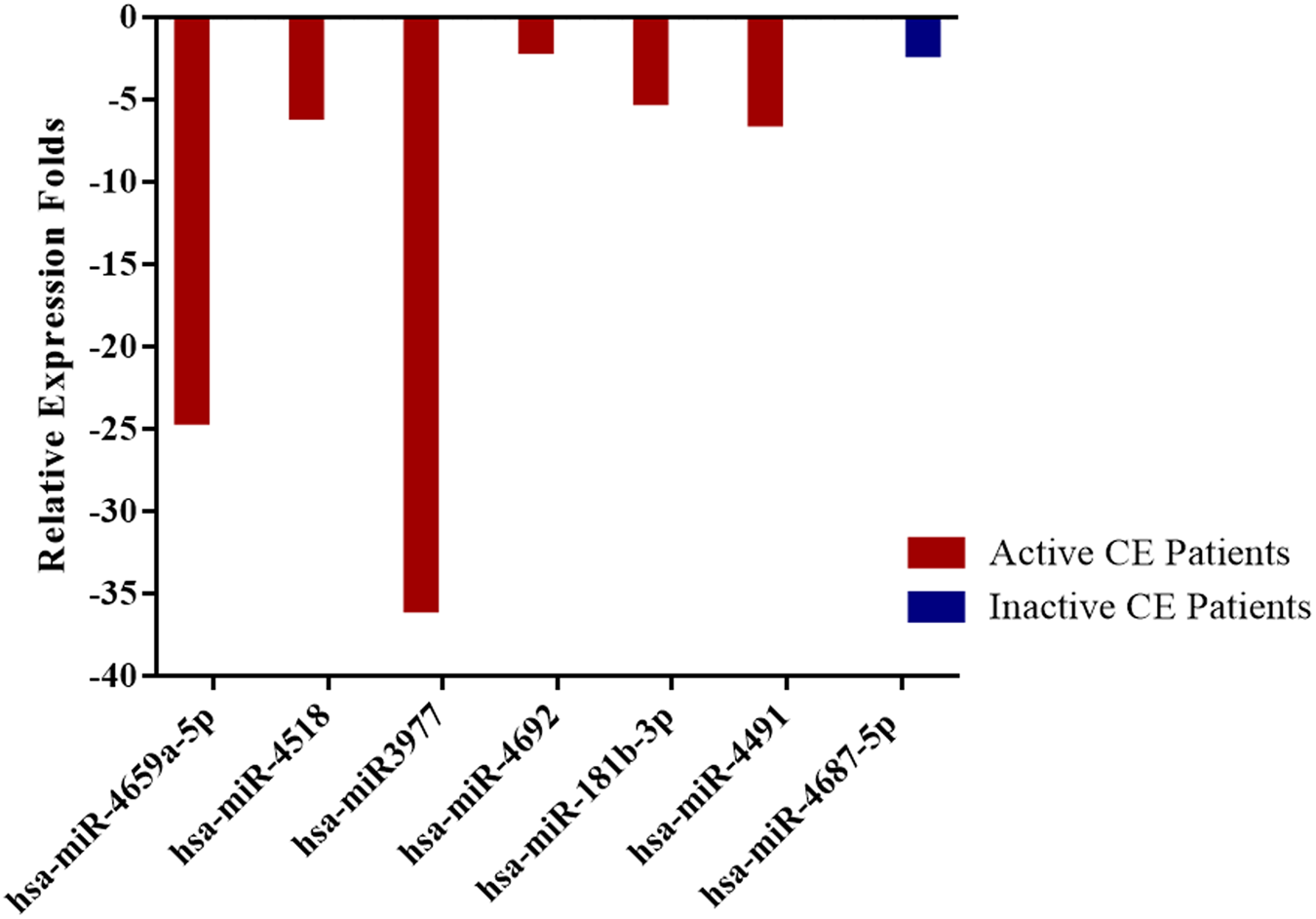
Fig. 2. Compared to control group downregulated miRNAs in active and inactive CE patients with a significant relative expression fold change (p < 0.05).
Pathway analysis
Downregulated miRNAs in active CE patients mainly regulate the expressions of cancer related oncogenes and tumour suppressor genes through cell proliferation, cell cycle, cell–cell interaction, transport systems, DNA repair and translational regulation. Additionally, these miRNAs can also play a role in neuronal growth process, function, differentiation and expression of immunoglobulin superfamilies.
Downregulated miRNA in inactive CE patients mainly regulates various cellular processes, including cell cycle progression, signal transduction, apoptosis, and gene regulation.
Discussion
Since the correlation between the expression of miRNAs in circulation and various pathologies has been determined, miRNAs are thought to be promising diagnostic biomarkers (Schwarzenbach et al., Reference Schwarzenbach, Nishida, Calin and Pantel2014). Although many studies focused on miRNA expression profiles in parasitic infections, there are only two studies conducted on the miRNA-based analysis of CE patients. In the study conducted by Mariconti et al., the expression profile of immune-related miRNAs was reported to be altered between active and inactive CE patients (Mariconti et al., Reference Mariconti, Vola, Manciulli, Genco, Lissandrin, Meroni, Rosenzvit, Tamarozzi and Brunetti2019). According to their findings, six miRNAs in active CE patients were found to be upregulated compared with inactive CE patients. miRNAs detected in their study, have role in various immune-related processes such as proliferation/activation of macrophages, inflammation, apoptosis and/or oxidative damage, the regulation of the innate immunity, the type I interferon signalling and tumour suppression in many types of cancers (Mariconti et al., Reference Mariconti, Vola, Manciulli, Genco, Lissandrin, Meroni, Rosenzvit, Tamarozzi and Brunetti2019). In the other study, parasite-derived miRNAs, egr-miR71 and egr-let 7, which were detected in circulation of CE patients found to be decreased after the removal of the cyst. These miRNAs are considered promising biomarkers for CE; however, the study does not provide any information about the pathways affected in the host in presence of CE (Alizadeh et al., Reference Alizadeh, Mahami-Oskouei, Spotin, Kazemi, Ahmadpour, Cai, Shanehbandi and Shekari2020).
According to our results, compared with control group downregulation in six miRNAs and one miRNA expressions were determined in active and inactive CE patients, respectively. These miRNAs were already known to be mainly involved in cell proliferation, apoptosis, cell–cell interactions and cell cycle. Current knowledge has suggested that miRNAs can play antitumoral or oncogenic roles in different cancer types. It is known that hsa-miR-4659a-5p, hsa-miR-4518, hsa-miR-181b-3p and hsa-miR-4687-5p, which were determined to be downregulated in this study, have a role in cancer development. In particular, an association between hsa-miR-4659a-5p and advanced breast cancer has been reported (Tabatabian et al., Reference Tabatabian, Mesrian Tanha, Tabatabaeian, Sadeghi, Ghaedi and Mohamadynejad2020). In a recent study, miR-4518 expression was found to be downregulated significantly in glioma tissues (Lu et al., Reference Lu, Cai and Li2018). miR-181b-3p was found to induce epithelial–mesenchymal transition which is crucial for increased invasion and metastasis during cancer progression in MCF7 breast cancer cells (Yoo et al., Reference Yoo, Kwak, An, Bae, Park and Han2016). Additionally, miR-181b-3p was recently found as one of the miRNAs with potential in discriminating neck lymph node metastasis, suggesting that this miRNA has a potential as a prognostic biomarker (Liu et al., Reference Liu, Zhu and Brooks2020). Lastly, miR-4687-5p was found to be upregulated in lung adenocarcinoma metastasis and considered to be among potential miRNA biomarkers for small cell lung cancer (Xu et al., Reference Xu, Wang, Sun and Xin2020).
Some parasitic infections are well-known risk factors for various types of cancer in mammalian hosts. On the other hand, reports on the anticancer effects of parasitic organisms are limited. However, there are conflicting reports on the effect of E. granulosus on cancer risk. A retrospective study on CE suggested a negative correlation between CE and solid tumours (Akgül et al., Reference Akgül, Tez, Unal, Keşkek, Sayek and Ozçelik2003). In contrast, Oikonomopoulou et al. proposed that infection by E. granulosus may increase the risk of cancer (Oikonomopoulou et al., Reference Oikonomopoulou, Yu and Wang2016). Relationship between CE and cancer has not been clearly defined yet. Many studies also suggest a protective effect of E. granulosus and its products against cancer development. However, molecular mechanisms still need to be clarified (Guan et al., Reference Guan, Zhang, Wang, Lu, Yin and Zhang2019).
To date, several mechanisms have been proposed to explain CE related anticancer effects, including parasite molecules and activation of host immune response. Ranasinghe et al. proposed that Kunitz-type protease inhibitor EgKI-1 which is highly expressed by the oncosphere of E. granulosus could disrupt the regular cell cycle and induce apoptosis in cancer cells (Ranasinghe et al., Reference Ranasinghe, Fischer, Zhang, Gobert and McManus2015, Reference Ranasinghe, Boyle, Fischer, Potriquet, Mulvenna and McManus2018).
In this study, most of the downregulated miRNAs in CE patients were found to be associated with cell proliferation, apoptosis, cell–cell interactions and cell cycle. Downregulation of these miRNAs due to the presence of the CE cyst possibly play a regulatory role in the anti-cancer effect.
This research has particular limitations. The patients in this study were limited to liver CE Thus, further research including CE patients with other organ involvement like lungs, kidneys, etc., is needed to verify whether these miRNAs play an essential role in E. granulosus infection.
In conclusion, we showed that profiles of miRNA expression vary in active and inactive CE cysts of the liver. According to pathway analysis based on alterations in miRNA profiles, targets were predicted to involve mainly cell proliferation, apoptosis, cell–cell interactions and cell cycle control. miRNAs downregulated in CE should be considered as promising molecules in the mechanism based on the relation between cancer development and CE. Further studies based on host miRNAs are needed to confirm the affected genes in the presence of CE and to enlighten underlying molecular mechanisms of the relationship between CE and cancer.
Financial support
This study was supported by Hacettepe University Scientific Research Projects Coordination Unit (Grant Number: TSA-2019-17873).
Conflicts of interest
None.





