Mastitis control is the most frequent reason for antibiotic use in lactating and non-lactating dairy cows (Pol and Ruegg, Reference Pol and Ruegg2007). Because of concerns that antibiotic usage may lead to antimicrobial resistance (WHO, 2015), strategies are needed to promote and ensure prudent use of antimicrobials for mastitis control. Selective treatment for CM in lactating cows, based on differentiation between gram-positive pathogens and other causes of mastitis, has the potential to reduce antibiotic use significantly without negative impact on udder health, production or culling (Lago et al., Reference Lago, Godden, Bey, Ruegg and Leslie2011a, Reference Lago, Godden, Bey, Ruegg and Leslie2011b; Mansion-de Vries et al., Reference Mansion-de Vries, Knorr, Paduch, Zinke, Hoedemaker and Krömker2014). This has led to development of an array of diagnostic tools for on-farm classification of mastitis pathogens to support selective treatment.
Numerous culture-based detection kits for classification of mastitis pathogens have been reviewed, and new tests are becoming commercially available (Malcata et al., Reference Malcata, Pepler, O'Reilly, Brady, Eckersall, Zadoks and Viora2020). There are diagnostic tests based on Petrifilm, agar plates, or tube-test based systems. Some identify bacteria as gram-positive or gram-negative, whereas other tests identify bacteria to genus or species-level (Malcata et al., Reference Malcata, Pepler, O'Reilly, Brady, Eckersall, Zadoks and Viora2020). Some assays also include antibiotic susceptibility testing (Jones et al., Reference Jones, Bork, Ferguson and Bates2019). All tests are more reliable when used for diagnosis of broad categories, such as growth, gram-positive and gram-negative species, rather than at genus or species level (Lago and Godden, Reference Lago and Godden2018). The performance of diagnostic assays can be evaluated using scientific characteristics such as sensitivity, specificity and accuracy, and convenience aspects such as cost, ease of use and turn-around time. For example, most Petrifilm or agar-based tests are cheap but require considerable user training whereas Mastatest (Jones et al., Reference Jones, Bork, Ferguson and Bates2019) costs more but provides automated reading to increase ease of use.
Although many tests were designed to identify pathogens to genus or species level, farmers are more interested in advice on antibiotic use than identification of causative agents of CM (Griffioen et al., Reference Griffioen, Hop, Holstege, Velthuis and Lam2016). A simplified test to differentiate gram-positive organisms from other causes of mastitis could be sufficient to decide whether antimicrobial treatment of non-severe CM is needed. In a different context, namely bacteriuria in pregnant women, a similar need for reliable and simple testing to differentiate gram-positive, gram-negative and culture-negatives samples led to development of the Uricult dip-slide (Van Dorsten and Bannister, Reference Van Dorsten and Bannister1986). The dip-slide is a plastic paddle with two selective media that can be dipped in a liquid sample such as urine or milk, allowing for growth of either gram-positive or gram-negative organisms.
The aim of our study was to evaluate the laboratory performance of a simplified slide test for bovine mastitis, to determine whether it differentiates gram-positive from other forms of mastitis with similar accuracy as a commercially available comparator test commonly used in our practice.
Materials and methods
Regulatory compliance
This research was approved by the Ethics and Welfare Committee, School of Veterinary Medicine, University of Glasgow, UK (Ref 50a/16).
Sample collection
Seven dairy farms in Scotland were selected based on herd size, location, and willingness to cooperate in the study (online Supplementary Table S1). Farm staff, including milkers and herd managers, were trained to identify CM and to classify cases as mild (abnormal milk, e.g. clots, flakes or serous milk), moderate (abnormal milk and signs of udder inflammation: hardness, swelling, redness, heat or pain) or severe (presence of additional systemic signs of disease, e.g. fever, tachycardia, tachypnoea, dehydration, or anorexia) (Pinzón-Sánchez and Ruegg, Reference Pinzón-Sánchez and Ruegg2011). They were taught how to collect milk samples aseptically according to National Mastitis Council recommendations (NMC, 2017). CM cases were sampled regardless of mastitis severity. If multiple quarters of a cow were affected simultaneously, each affected quarter was sampled. Any CM episode in a quarter occurring >14 d after the previous episode, or caused by a different aetiological agent, was considered a new CM case. Animals were eligible for inclusion in the first week after calving but no animals included were within 14 d of administration of antimicrobial products.
Samples were collected from January to May 2018. They were stored on farm at −20°C, and transported once a week to Glasgow University's Veterinary Diagnostic Services laboratory where they were stored at −20°C until processing. All samples were cultured within 4 weeks from CM detection.
Reference test
Samples were thawed at ambient temperature for up to 8 h and processed simultaneously using the reference test, the simplified slide test, and the commercially available plate-based comparator test as described in the following two sections. For consistency, all media were inoculated and read by the first author, starting with the slide test. Bacteriological culture (NMC, 2017) with subsequent determination of species identity using MALDI-ToF MS was used as the reference test as detailed in the supplementary file.
Slide test
Media of the simplified slide test (VétoSlide, Vétoquinol, Lure, France) were inoculated by applying milk directly to each side using cotton wool swabs (approximate volume 0.1 ml) to moisten the entire surface of the media, as per manufacturer's instructions. The inoculated slides were incubated aerobically at 37°C and examined after 24–48 h. When at least one colony was visible, the sample was considered positive (Dohoo et al., Reference Dohoo, Smith, Andersen, Kelton and Godden2011). Based on the manufacturer's guidelines, any growth on the green media was considered to indicate presence of gram-negative bacteria and red colonies on the green media were considered Escherichia coli. Growth on the red media was considered to indicate presence of gram-positive bacteria (online Supplementary Figure S1). When there was growth on both media, it was considered to indicate mixed infection with gram-positive and gram-negative bacteria. Guidelines to identify contaminated samples were not given, so samples were never classed as contaminated based on the slide test.
Comparator test
Plates for the comparator test (VétoRapid, Vétoquinol, Lure, France) were chosen for benchmarking because it is the most commonly used on-farm test in the dairy community of the authors and was previously evaluated in similar study settings (Viora et al., Reference Viora, Graham, Mellor, Reynolds, Simoes and Geraghty2014). These plates were inoculated with 0.01 ml of milk per sector using disposable sterile calibrated plastic loops, incubated aerobically at 37°C and examined after 24–48 h, as detailed in the supplementary file. Results were summarized as gram-positive, gram-negative, E. coli and no growth for comparison with the reference and slide tests results. Samples not yielding visible colonies on the comparator test were considered negative for mastitis-associated pathogens. As for the slide test, a contaminated category was not specified by the manufacturer.
Data analysis
Samples that were contaminated or contained non-identifiable isolates by MALDI-ToF MS were excluded from evaluation of diagnostic test performance. All other culture-positive and culture-negative samples (n = 130) were used to calculate sensitivity, specificity, accuracy, positive predictive values (PPV) and negative predictive values (NPV) for growth, gram-positives, gram-negatives and E. coli. The reference test was used to classify results from the slide test and comparator test as correct or incorrect. To evaluate the potential of the test kits as treatment decision support tools, the calculations were repeated using a subset of the 130 samples, namely those from non-severe CM cases (n = 109), and the outcome was expressed as ‘treatment’. This outcome is equivalent to gram-positive growth or no gram-positive growth. The latter category includes gram-negative bacteria, non-bacterial growth, and culture-negative results.
Statistical analysis was performed in Excel (Microsoft Corp., Redmond, USA) using tabular methods, and in R. If the 95% confidence interval for the difference between tests excluded zero, test performance was considered significantly different. Full details are provided in the supplementary file.
Results
Reference test
Between 5 and 58 samples were collected from each farm (online Supplementary Table S1). Of 156 samples, 23 (14.7%) were contaminated. Among 133 non-contaminated samples, 14 (10.5%) showed no growth, and 116 (87.2%) showed growth of one or two colony types that could be identified by the reference method (Table 1). Three samples with growth of organisms that could not be identified by the reference method were excluded from further analysis. Within each farm's sample set, gram-positive and gram-negative isolates were identified, with a preponderance of gram-positive results for some farms (Farms 3, 4 and 5), mostly gram-negative results for others (Farms 2 and 6) and an even balance for the remainder (Farms 1 and 7; Figure 1). The proportion of contaminated samples per farm ranged from 0 to 33%, indicating considerable differences in sample quality. The most common species were E. coli and Streptococcus uberis, followed by other major mastitis pathogens, including Streptococcus dysgalactiae, Staphylococcus aureus and Klebsiella spp. (Table 1).
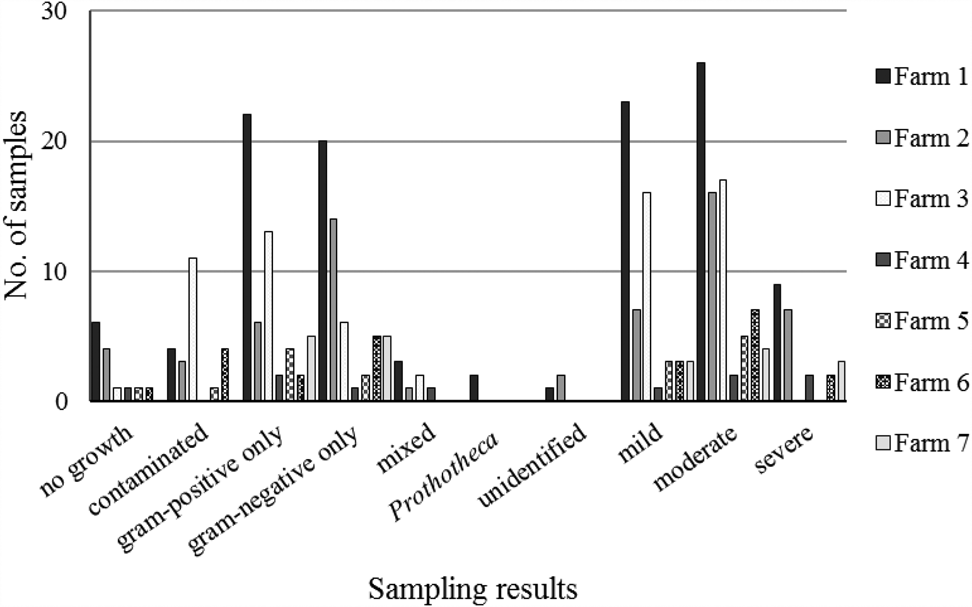
Fig. 1. Sampling results for bovine milk samples (n = 130) from quarters with clinical mastitis by participating farm. The number of samples for each farm were, from farm 1 to 7 respectively, 58, 30, 35, 5, 8, 12 and 10.
Table 1. Test results of 130 milk samples from bovine clinical mastitis based on a reference test consisting of standard bacteriological culture and species identification by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-ToF MS) in comparison with the simplified slide test under evaluation (VétoSlide, Vétoquinol, Lure, France) and a commercially available plate-based comparator (VétoRapid, Vétoquinol, Lure, France)
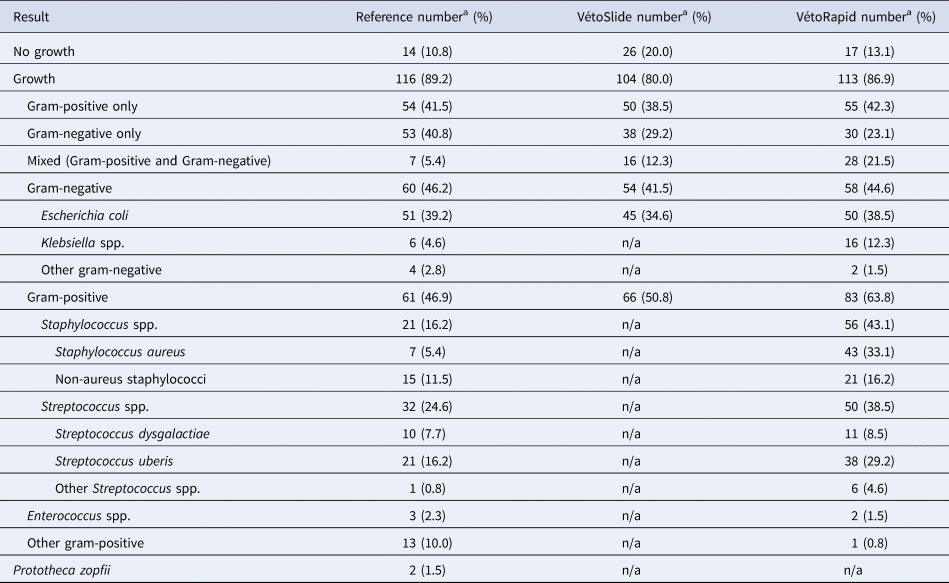
n/a, not applicable.
a The total number of species/genera listed exceeds the number of samples because more than one species/genus was detected in some samples that were not contaminated based on the NMC standard definition of 3 or more colony types (the percentage shown is related to the proportion of samples).
Slide test
After excluding contaminated samples and those with unidentified organisms, 130 samples were used to evaluate the performance of the slide test. A milk sample could be culture negative or culture positive, contain a single colony type or two colony types (two gram- positive morphotypes, two gram-negative morphotypes, or mixed gram-positive and gram-negative growth). The latter were considered gram-positive in the gram-positive analysis and gram-negative in the gram-negative analysis. The proportion of culture negative results was considerably higher for the slide test (20%) than for the reference test (10.8%). Of 26 culture-negative samples in the slide test, 12 (46.2%) were correctly classified. Of 14 false negative slide test results, seven were from samples with gram-positive growth in the reference test and seven from samples with gram-negative growth.
Mixed gram-positive and gram-negative growth was more common in the slide test (12.3%) than in the reference test (5.4%) (Table 1). The sensitivity of the slide test was similar for gram-positive and gram-negative organisms, but specificity was higher for the latter, resulting in higher accuracy for gram-negative organisms (89.2%) or E. coli (92.3%) than for gram-positive organisms (79.2%: Table 2). For the samples from non-severe CM (n = 109), the potential of the slide test to be used as treatment decision support tool was evaluated. Sensitivity and specificity for this subset were similar to those for all CM cases (Table 2).
Table 2. Performance of the simplified slide test under evaluation (VétoSlide, Vétoquinol, Lure, France) and a commercially available plate-based comparator (VétoRapid, Vétoquinol, Lure, France) for identification of mastitis pathogens (n = 130 samples) and as a treatment decision support tools for non-severe clinical mastitis (n = 109 samples)
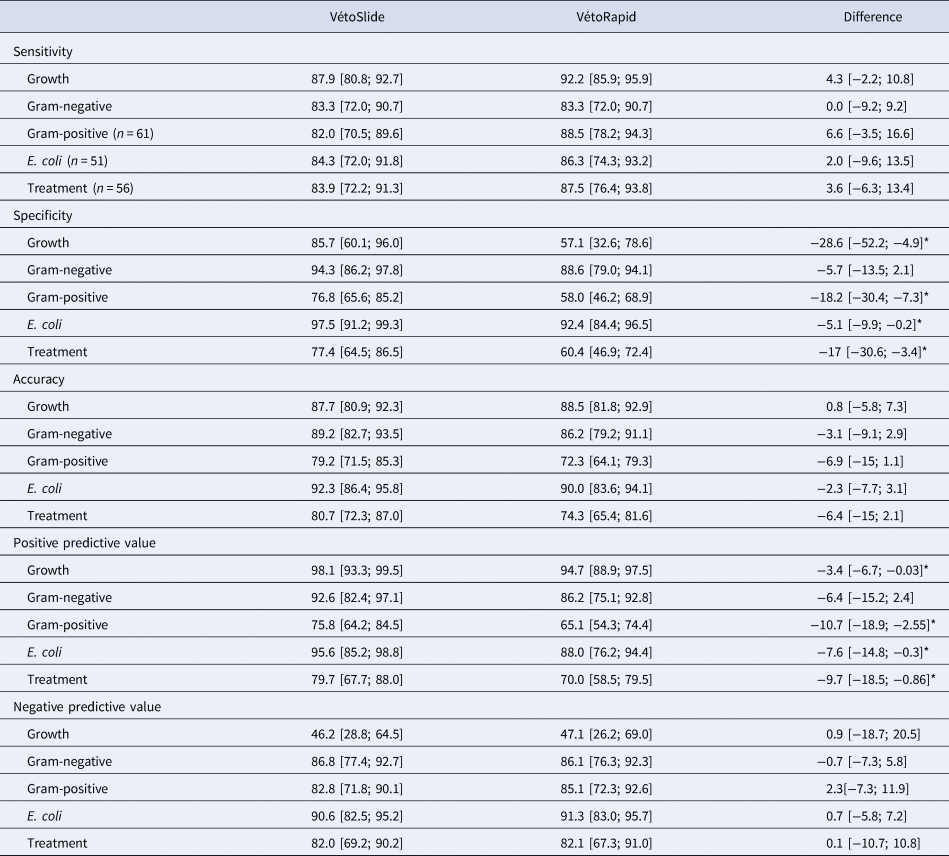
*Outcomes where VétoSlide and VétoRapid are significantly different.
The total of samples that yield in the reference test growth, Gram-negative, Gram-positive, E. coli and Treatment were, 116, 60, 61, 51 and 56 respectively.
Results are based on comparison with a reference test consisting of culture and species identification based on matrix-assisted laser-desorption ionization time-of-flight mass spectrometry and the difference compares the two tests while correcting for dependence. Values are point estimates expressed as percentages, with 95% confidence intervals in brackets.
Mean PPV of the slide test was high (between 92.6 and 98.1%) for growth, gram-negative results, and E. coli and moderate (between 75 and 80%) for gram-positive results (all based on 130 samples) and treatment (based on 109 samples). The mean NPV was less than 50% for growth and between 82 and 90.6% for all other outcomes (Table 2).
Benchmarking against the comparator test
Despite using a higher inoculum, the slide test gave fewer false positive results for growth than the comparator test, resulting in significantly higher specificity. Low specificity of the comparator test was partly due to moderate specificity in the detection of Staph. aureus and Strep. uberis (70.7 and 82.6%, respectively; online Supplementary Table S2), which was accompanied by high sensitivity for the same pathogens (100 and 90.5%, respectively). For sensitivity, there were no statistically significant differences among tests for any of the outcomes (Table 2). Despite the greater specificity of the slide test, overall accuracy of the two tests was not significantly different. In our study population, the slide test had significantly greater PPV than the comparator test for growth, gram-positive, E. coli and treatment, whereas their NPVs were similar for all outcomes.
Discussion
We evaluated the laboratory performance of a simplified culture-based slide test, VétoSlide, which was developed as a potential point-of-care tool to support farmers' CM treatment decisions. Its accuracy for gram-positive organisms in samples from non-severe CM (80.7%) is in the same range as commercially available point-of-care tests, including the comparator test (VétoRapid) (74.3%, this study), the Minnesota Easy Culture System-Triplate (81.3%) (Ferreira et al., Reference Ferreira, Gomes, Bonsaglia, Canisso, Garrett, Stewart, Zhou and Lima2018), Minnesota Easy Culture System-biplate (81 to 84%) (Royster et al., Reference Royster, Godden, Goulart, Dahlke, Rapnicki and Timmerman2014), Petrifilm (80.2%) (Mansion-de Vries et al., Reference Mansion-de Vries, Knorr, Paduch, Zinke, Hoedemaker and Krömker2014) and MastDecide (58.6 to 85.3%) (Leimbach and Krömker, Reference Leimbach and Krömker2018), although comparisons between studies are complicated by differences in study design, populations, and methods of analysis. Such differences make comparison of predictive values problematic because they are highly dependent on pathogen prevalence, which is farm-specific. Benchmarking of the new slide test against a commercially available comparator in a single study allowed us to compare predictive values, which are more important in practice than sensitivity, specificity, or accuracy. A high PPV means that unnecessary treatment is minimized whereas a high NPV means that treatment is withheld only when cows truly do not need it. The slide test outperformed the comparator test in PPV and had similar NPV, meaning that the reduced risk of over-treating was not accompanied by an increased risk of under-treating. Whether positive or negative predictive value is considered more important in informing treatment decisions differs between regions. In some areas, such as southern Europe, it is generally assumed that antimicrobial treatment of mastitis is needed until proven otherwise (Busani et al., Reference Busani, Graziani, Franco, Di Egidio, Binkin and Battisti2004). Conversely, in northern Europe, it is assumed that treatment is not needed until proven otherwise (Persson Waller et al., Reference Persson Waller, Hardemark, Nyman and Duse2016). Within countries, this balance may shift over time, as illustrated by work from The Netherlands on selective dry cow treatment (DCT). Two split-udder trials conducted two decades apart (Schukken et al., Reference Schukken, Vanvliet, Vandegeer and Grommers1993; Scherpenzeel et al., Reference Scherpenzeel, den Uijl, van Schaik, Olde Riekerink, Keurentjes and Lam2014) in the same country both showed that blanket DCT prevents CM when compared to selective DCT. However, the first study concluded that blanket DCT should be used to prevent CM despite the need to ‘eliminate unnecessary use of antibiotics’, whereas the second study emphasized the reduction in antimicrobial use that could be achieved by abandoning blanket DCT.
Whether the price of the diagnostic test is worth paying in terms of financial benefit is a matter of debate. On farms with a single dominant pathogen, the value of information may be limited (Cha et al., Reference Cha, Smith, Kristensen, Hertl, Schukken, Tauer, Welcome and Gröhn2016), but our data showed that several farms did not have clear predominance of gram-positive or gram-negative mastitis over other types of mastitis. Some authors argue that even with just 20% of gram-positive mastitis, use of on-farm diagnostics would not be cost-effective (Down et al., Reference Down, Bradley, Breen and Green2017). Hence, the value of information would be farm-specific and no blanket statements around cost-benefit or reductions in antimicrobial use can be made based on our results. It is clear, however, that some farms will need further training in sample collection and handling to reduce the number of contaminated samples and to make investment of time and money into diagnostic testing better value for money. Moreover, before uptake of the slide test can be recommended on-farm, evaluation under on-farm conditions will be needed, as our laboratory-based analysis included freezing and thawing of milk, which would not be part of its on-farm use. Reading of plates at 24 and 48 h, as done here to allow for comparison with VétoRapid results, would cause considerable delay in treatment decisions, and shorter incubation times would need to be considered, with growth of gram-negative organisms often visible well within 24 h (data not shown).
The reference test used in our study included species identification by MALDI-ToF MS and revealed the presence of several species that are not recognized as typical mastitis pathogens, e.g. Bacillus and Lysinibacillus species. Although both genera are gram-positive, it is debatable whether they should be targeted with antimicrobial treatment because little is known about their role as pathogenic agents or their response to treatment. None of the currently available point-of-care tests for mastitis have the ability to differentiate such organisms from recognized mastitis pathogens. When information at species or subspecies level is required for advanced investigations or decision making at herd or animal level, laboratory-based microbial diagnostics continue to be important (Mansion-de Vries et al., Reference Mansion-de Vries, Knorr, Paduch, Zinke, Hoedemaker and Krömker2014). For on-farm treatment decision making, however, the simplified slide test appears to have the potential to be an affordable, accurate, and user-friendly option.
In conclusion, using laboratory-based evaluation of farmer collected milk samples we demonstrated that a simplified slide test performs similar to the commercially available on-farm test that was used for benchmarking in terms of sensitivity or accuracy, whilst performing better in terms of specificity. The simplicity of the slide test can make it an attractive tool for farmers to target antimicrobial treatment of non-severe CM cases caused by gram-positive organisms with good diagnostic accuracy. Further evaluation of user-friendliness and test accuracy in on-farm settings is needed, followed by assessment of uptake, economic impact, and reduction in antimicrobial use. In addition, users' willingness and ability to collect high quality milk samples needs to be understood and supported.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0022029921000303
Acknowledgements
We would like to thank the staff at the participating farms, our colleagues and the Veterinary Diagnostic Services, University of Glasgow, for their time and effort. Financial and in-kind support for this project was received from Vétoquinol, Lure, France.





