No CrossRef data available.
Published online by Cambridge University Press: 06 November 2024
This study was conducted to estimate the relative contribution of dominance genetic effects to efficiency-related traits including Kleiber ratio (KR), efficiency of growth (EF) and relative growth rate (RGR) in Baluchi sheep. To this end, each trait was analysed with a series of 12 animal models which were identical for fixed and additive genetic effects but differed for combinations of dominance genetic and maternal effects. The Akaike's information criterion (AIC) was used to rank models. (Co)variances between traits were estimated using bivariate analyses. For all traits studied, according to AIC values, models containing the dominance genetic effects provided a better data fit than otherwise identical models. By including dominance genetic effects in the model, additive genetic variance did not change, but a significant decrease was observed in the residual variance (24, 19 and 25% for KR, EF and RGR, respectively). Estimates of dominance heritability $( {\boldsymbol h}_{\boldsymbol d}^ 2 )$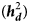 were 0.20 ± 0.05, 0.17 ± 0.05 and 0.19 ± 0.07 for KR, EF and RGR, respectively, more than corresponding estimates of additive heritability ${\bf ( }{\boldsymbol h}_{\boldsymbol a}^{\bf 2} {\bf ) }$
were 0.20 ± 0.05, 0.17 ± 0.05 and 0.19 ± 0.07 for KR, EF and RGR, respectively, more than corresponding estimates of additive heritability ${\bf ( }{\boldsymbol h}_{\boldsymbol a}^{\bf 2} {\bf ) }$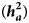 as 0.14 ± 0.02, 0.09 ± 0.03 and 0.13 ± 0.02, respectively. Dominance genetic correlations between traits were 0.89 ± 0.17 (KR-EF), 0.86 ± 0.20 (KR-RGR) and 0.93 ± 0.21 (EF-RGR). Additive genetic correlations between traits were 0.84 ± 0.05 (KR-EF), 0.78 ± 0.04 (KR-RGR) and 0.83 ± 0.04 (EF-RGR). The Spearman correlation between additive breeding values including and excluding dominance genetic effects were close to unity either for all animals or top ranked animals. Since presence of dominance genetic effects increased the model power to fit the data, inclusion of these effects in the genetic evaluation models for Baluchi sheep was recommended.
as 0.14 ± 0.02, 0.09 ± 0.03 and 0.13 ± 0.02, respectively. Dominance genetic correlations between traits were 0.89 ± 0.17 (KR-EF), 0.86 ± 0.20 (KR-RGR) and 0.93 ± 0.21 (EF-RGR). Additive genetic correlations between traits were 0.84 ± 0.05 (KR-EF), 0.78 ± 0.04 (KR-RGR) and 0.83 ± 0.04 (EF-RGR). The Spearman correlation between additive breeding values including and excluding dominance genetic effects were close to unity either for all animals or top ranked animals. Since presence of dominance genetic effects increased the model power to fit the data, inclusion of these effects in the genetic evaluation models for Baluchi sheep was recommended.