Crossref Citations
This article has been cited by the following publications. This list is generated based on data provided by
Crossref.
Tappiban, Piengtawan
Smith, Duncan R.
Triwitayakorn, Kanokporn
and
Bao, Jinsong
2019.
Recent understanding of starch biosynthesis in cassava for quality improvement: A review.
Trends in Food Science & Technology,
Vol. 83,
Issue. ,
p.
167.
Torres, Lívia Gomes
Vilela de Resende, Marcos Deon
Azevedo, Camila Ferreira
Fonseca e Silva, Fabyano
de Oliveira, Eder Jorge
and
Zhang, Peng
2019.
Genomic selection for productive traits in biparental cassava breeding populations.
PLOS ONE,
Vol. 14,
Issue. 7,
p.
e0220245.
do Carmo, Cátia Dias
e Sousa, Massaine Bandeira
Brito, Ana Carla
and
de Oliveira, Eder Jorge
2020.
Genome-wide association studies for waxy starch in cassava.
Euphytica,
Vol. 216,
Issue. 5,
Gimase, James M.
Thagana, Wilson M.
Omondi, Chripine O.
Cheserek, Jane J.
Gichimu, Bernard M.
Gichuru, Elijah K.
Ziyomo, Cathrine
and
Sneller, Clay H.
2020.
Genome-Wide Association Study identify the genetic loci conferring resistance to Coffee Berry Disease (Colletotrichum kahawae) in Coffea arabica var. Rume Sudan.
Euphytica,
Vol. 216,
Issue. 6,
dos Santos Silva, Priscila Patrícia
Sousa, Massaine Bandeira e
de Oliveira, Eder Jorge
Morgante, Carolina Vianna
de Oliveira, Carlos Roberto Silva
Vieira, Simone Leal
and
Borel, Jerônimo Constantino
2021.
Genome-wide association study of drought tolerance in cassava.
Euphytica,
Vol. 217,
Issue. 4,
Torres, Lívia Gomes
Oliveira, Eder Jorge de
Ogbonna, Alex C.
Bauchet, Guillaume J.
Mueller, Lukas A.
Azevedo, Camila Ferreira
Fonseca e Silva, Fabyano
Simiqueli, Guilherme Ferreira
and
Resende, Marcos Deon Vilela de
2021.
Can Cross-Country Genomic Predictions Be a Reasonable Strategy to Support Germplasm Exchange? – A Case Study With Hydrogen Cyanide in Cassava.
Frontiers in Plant Science,
Vol. 12,
Issue. ,
Mbanjo, Edwige Gaby Nkouaya
Rabbi, Ismail Yusuf
Ferguson, Morag Elizabeth
Kayondo, Siraj Ismail
Eng, Ng Hwa
Tripathi, Leena
Kulakow, Peter
and
Egesi, Chiedozie
2021.
Technological Innovations for Improving Cassava Production in Sub-Saharan Africa.
Frontiers in Genetics,
Vol. 11,
Issue. ,
Callegari, Daihany Moraes
Lima, Aline Medeiros
Ferreira Barros, Nicolle Louise
Siqueira, Andrei Santos
Moura, Elisa Ferreira
and
Batista de Souza, Cláudia Regina
2021.
Changes in transcript levels of cassava superoxide dismutase and catalase during interaction with Phytopythium sp..
Physiological and Molecular Plant Pathology,
Vol. 114,
Issue. ,
p.
101629.
Hohenfeld, Camila Santiago
Passos, Adriana Rodrigues
de Carvalho, Hélio Wilson Lemos
de Oliveira, Saulo Alves Santos
de Oliveira, Eder Jorge
and
Adhimoolam, Karthikeyan
2022.
Genome-wide association study and selection for field resistance to cassava root rot disease and productive traits.
PLOS ONE,
Vol. 17,
Issue. 6,
p.
e0270020.
Sholihin
Widodo
Susanawati
Senge, M.
Aziz, A.A.
Witono, Y.
Sharifuddin, J.
Robani, A.B.
Krisnamurthi, B.
Saiyut, P.
Mulyo, J.H.
bin Kamarudin, M.F.
and
Tjale, M.M.
2022.
Vamas 1, a new early root bulking, high-yielding, high-starch content cassava variety.
E3S Web of Conferences,
Vol. 361,
Issue. ,
p.
04014.
Li, Ruimei
Yuan, Shuai
Zhou, Yangjiao
Wang, Shijia
Zhou, Qin
Ding, Zhongping
Wang, Yajie
Yao, Yuan
Liu, Jiao
and
Guo, Jianchun
2022.
Comparative Transcriptome Profiling of Cassava Tuberous Roots in Response to Postharvest Physiological Deterioration.
International Journal of Molecular Sciences,
Vol. 24,
Issue. 1,
p.
246.
Santos, Cristiano Silva dos
Sousa, Massaine Bandeira
Brito, Ana Carla
de Oliveira, Luciana Alves
Carvalho, Carlos Wanderlei Piler
de Oliveira, Eder Jorge
and
Raman, Harsh
2022.
Genome-wide association study of cassava starch paste properties.
PLOS ONE,
Vol. 17,
Issue. 1,
p.
e0262888.
Paliwal, Rajneesh
and
Abberton, Michael
2022.
Next-Generation Sequencing and Agriculture.
p.
92.
Ghasemi, M.
Ghafouri-Kesbi, F.
and
Zamani, P.
2023.
Genomic evaluation of threshold traits in different scenarios of threshold number using parametric and non-parametric statistical methods.
The Journal of Agricultural Science,
Vol. 161,
Issue. 1,
p.
109.
Alleyne, Angela
Mason, Shanice
and
Vallès, Yvonne
2023.
Characterization of the Cassava Mycobiome in Symptomatic Leaf Tissues Displaying Cassava Superelongation Disease.
Journal of Fungi,
Vol. 9,
Issue. 12,
p.
1130.
Hohenfeld, Camila Santiago
de Oliveira, Saulo Alves Santos
Ferreira, Claudia Fortes
Mello, Victor Hugo
Margarido, Gabriel Rodrigues Alves
Passos, Adriana Rodrigues
and
de Oliveira, Eder Jorge
2024.
Comparative analysis of infected cassava root transcriptomics reveals candidate genes for root rot disease resistance.
Scientific Reports,
Vol. 14,
Issue. 1,
Sunvittayakul, Pongsakorn
Wonnapinij, Passorn
Chanchay, Pornchanan
Wannitikul, Pitchaporn
Sathitnaitham, Sukhita
Phanthanong, Phongnapha
Changwitchukarn, Kanokpoo
Suttangkakul, Anongpat
Ceballos, Hernan
Gomez, Leonardo D.
Kittipadakul, Piya
and
Vuttipongchaikij, Supachai
2024.
Genome-Wide Association Studies of Three-Dimensional (3D) Cassava Root Crowns and Agronomic Traits Using Partially Inbred Populations.
Agronomy,
Vol. 14,
Issue. 3,
p.
591.
Sunvittayakul, Pongsakorn
Wonnapinij, Passorn
Wannitikul, Pitchaporn
Phanthanong, Phongnapha
Changwitchukarn, Kanokpoo
Suttangkakul, Anongpat
Utthiya, Supanut
Phraemuang, Apimon
Kongsil, Pasajee
Prommarit, Kamonchat
Ceballos, Hernan
Gomez, Leonardo D.
Kittipadakul, Piya
and
Vuttipongchaikij, Supachai
2025.
Genome-wide association studies unveils the genetic basis of cell wall composition and saccharification of cassava pulp.
Plant Physiology and Biochemistry,
Vol. 218,
Issue. ,
p.
109312.
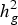 $h_g^2 $) was of medium magnitude for all groups of resistances for pathogens, with variation from 0·16 ± 0·019 (Fspp Pulp) to 0·31 ± 0·028 (Fspp Peel). The kinship matrix was used to correct for stratification as well as for clustering the accessions. Overall, this analysis showed that there was no relationship between agronomic traits and resistance to CRR and the four clusters obtained from kinship matrix. The GWAS identified 38 significant SNPs, of which eight and 22 are related to the severity of DRR in the pulp and peel, respectively. The other eight SNPs were associated with SRR-peel (1), SRR-pulp (1), BRR-peel (3) and BRR-pulp (3). Half of the SNPs associated with CRR resistance have functional annotations related to defence and response to pathogen attack as well as general cellular processes. The current study revealed that resistance to CRR is controlled by multiple loci with small effects, and significant SNPs can be used to identify putative genes that control these traits.
$h_g^2 $) was of medium magnitude for all groups of resistances for pathogens, with variation from 0·16 ± 0·019 (Fspp Pulp) to 0·31 ± 0·028 (Fspp Peel). The kinship matrix was used to correct for stratification as well as for clustering the accessions. Overall, this analysis showed that there was no relationship between agronomic traits and resistance to CRR and the four clusters obtained from kinship matrix. The GWAS identified 38 significant SNPs, of which eight and 22 are related to the severity of DRR in the pulp and peel, respectively. The other eight SNPs were associated with SRR-peel (1), SRR-pulp (1), BRR-peel (3) and BRR-pulp (3). Half of the SNPs associated with CRR resistance have functional annotations related to defence and response to pathogen attack as well as general cellular processes. The current study revealed that resistance to CRR is controlled by multiple loci with small effects, and significant SNPs can be used to identify putative genes that control these traits.


