Introduction
One objective of astrobiologists is to understand the habitability of extraterrestrial environments and to be able to investigate them for the presence of life or habitable conditions (Kasting & Catling Reference Kasting and Catling2003; Gaidos et al. Reference Gaidos, Deschenes, Dundon, Fagan, Menviel-Hessler, Moskovitz and Workman2005; Nisbet et al. Reference Nisbet, Zahnle, Gerasimov, Helbert, Jaumann, Hofmann, Benzerera and Westall2007; Southam et al. Reference Southam, Rothschild and Westall2007; Zahnle et al. Reference Zahnle, Arndt, Cockell, Halliday, Nisbet, Selsis and Sleep2007; Lammer et al. Reference Lammer2009; Stoker et al. Reference Stoker2010; Westall et al. Reference Westall, Loizeau, Foucher, Bost, Betrand, Vago and Kminek2013; Grotzinger et al. Reference Grotzinger2014; Cockell et al. Reference Cockell2016). To do this we need to know the limits of terrestrial life. ‘Maps’ of the limits of life can be used to investigate to what extent extraterrestrial environments fall within the known physical and chemical parameter space of the growth and reproduction of terrestrial life (Nichols et al. Reference Nichols, Greenhill, Shadbolt, Ross and McMeekin1999; Canganella & Wiegel Reference Canganella and Wiegel2011; Harrison et al. Reference Harrison, Gheeraert, Tsigelnitskiy and Cockell2013; Moissl-Eichinger et al. Reference Moissl-Eichinger, Cockell and Rettberg2016).
Investigating the limits of life requires that we have access to organisms from analogue environments that in some way share chemical and physical similarities to extraterrestrial conditions, so that the physiological and biochemical capabilities and limits of the organisms can be studied. A large number of analogue environments have been investigated, from deserts to inland lakes and deep subsurface environments (Smith et al. Reference Smith, Davis-Street, Fesperman, Smith, Rice and Zwart2004; Osinski et al. Reference Osinski, Léveillé, Berinstain, Lebeuf and Bamsey2006; Cabrol et al. Reference Cabrol2007; Skelley et al. Reference Skelley, Aubrey, Willis, Amashukeli, Ehrenfreund, Bada, Grunthaner and Mathies2007; Lim et al. Reference Lim, Garry and Bleacher2011; Abercromby et al. Reference Abercromby, Chappell and Gernhardt2013; Preston & Dartnell Reference Preston and Dartnell2014; Payler et al. Reference Payler2016). The focus of these studies covers a wide range from microbiology to instrument testing.
One set of terrestrial environments that have been less well studied with respect to determining the limits and locations of habitable extraterrestrial environments are anoxic analogue environments. Yet, most known extraterrestrial environments are oxygen-free or contain very low abundances of this gas. For example, the thin Martian atmosphere contains 0.14% oxygen and the surface and subsurface of that planet is therefore expected to be only suitable for microorganisms capable of growing under anaerobic conditions. Methods to simplify enriching and isolating anaerobic microorganisms have been developed over a long period of time (e.g. M'Leod Reference M'Leod1912; Kneteman Reference Kneteman1957; Hungate Reference Hungate, Norris and Robbins1969; Miller & Wolin Reference Miller and Wolin1974; Balch et al. Reference Balch, Fox, Magrum, Woese and Wolfe1979; Behbehani et al. Reference Behbehani, Jordan and Santoro1982; La Scola et al. Reference La Scola, Khelaifia, Lagier and Raoult2014), but nevertheless they remain challenging since anaerobic organisms are fastidious and usually grow much more slowly than aerobic organisms. This may explain why although many anaerobic isolates have been obtained over the years from diverse environments, isolating such microorganisms from analogue environments specifically is less commonly attempted than aerobic organisms.
Obtaining anaerobic organisms from analogue environments should ideally conform to several steps. Firstly, samples should be obtained under strict anoxic sampling methods that minimize exposure to oxygen. Secondly, organisms should be isolated using a defined medium, which can be easily reproduced. In terms of fundamental chemical recipe, it can also be the same for all enrichments so that the different organisms obtained can later be grown under identical medium conditions to allow their physiology and biochemistry to be compared. Different electron donors and acceptors can then be introduced into this basal medium. Thirdly, the obtained isolates should be deposited in an international culture collection so that they can be made available to other researchers, avoiding the laborious and usually haphazard process of obtaining them directly from diverse researchers.
In this paper, we describe systematic methods used by our consortium for the European Commission's Mars Analogues for Space Exploration (MASE) project and the results of the approach. The project spans from the selection of anoxic analogue field sites, the collection of anoxic samples for the characterization of the whole anaerobic community by sequencing, the enrichment and isolation of anaerobic organisms, their deposition within a culture collection for long-term storage as a pre-requisite for studies in physiology, biochemistry and other investigations.
Methods
Selection of field sites
A large number of field sites have been investigated as analogue sites. For the procedure developed here, we chose analogue sites based on three primary criteria: (1) the field sites should represent a collection of diverse geochemical conditions and physical/chemical extremes that provide a selection of putative early and present-day Mars aqueous environments; (2) some preliminary data should be available that suggests that they would yield useful samples for the study; and (3) they should be logistically easy to access and have accessible sampling locations from which anoxic samples could be easily collected using the proposed methodologies.
Collection of anoxic samples and sampling list
Prior to the sampling trip, glass bottles with an anoxic atmosphere were prepared in the laboratory. A summary of the materials used is given in Table 1. Half an ml of resazurin solution (0.1%, w/v), a redox-sensitive dye to monitor the redox potential of the introduced sample was pipetted into each 100 ml Duran glass bottle. The bottles were closed with a butyl rubber stopper, which was held in place with an appropriate screw cap containing an aperture. This step was followed by three gas–vacuum exchange cycles using nitrogen in order to obtain an oxygen-free atmosphere within the bottle. Finally, the glass bottle was purged with nitrogen until an overpressure of 0.2–0.5 bar was reached and it was autoclaved for 40 min at 121°C.
Table 1. Required components for obtaining anaerobic samples

Sampling procedure
In the field site, a specific protocol was followed for sample collection. Prior to removal of the screw cap and the rubber stopper, the overpressure within the anoxic glass bottle was released using a sterile needle. To avoid contamination, the butyl rubber stopper was wiped with ethanol-soaked wipes before injecting the needle into the rubber stopper. All steps within this protocol were performed wearing laboratory disposable gloves. Sediment samples were taken from the environment (lakes, pools and soils) as deep as possible (approximately 30–40 cm depth) to collect anoxic samples. Samples can be collected in a number of ways. In our case, we used a sterile 50 ml Falcon tube attached to the end of a pole, which improved sampling in soft sediments. In order to collect the sediment, the sampling tool was scooped through the sediment (for a distance of approximately 50 cm and a similar depth). Otherwise, the anoxic bottle could be opened under water and the sample could be collected by scooping through the sediment, filling the bottle with water up to the rim.
After filling the bottles, the bottles were closed with the rubber stopper. The utmost care is taken to avoid contamination and to minimize oxygen influx by air bubbles that remain in the glass bottle. An additional needle can be used to release excess liquid, while the rubber stopper was fitted to the glass bottle.
Sample storage
Following sampling, the samples were stored at 4°C under periodic observation to monitor the anoxic status of the samples by checking the colour. The resazurin dye provides an indication of the redox status in the sample. If the sample turns pink, then this indicates an atmosphere containing oxygen. One ml anoxic cysteine–HCl solution (2.5%, w/v) can be added to restore and maintain an anoxic atmosphere. Alternative reducing agents are sodium dithionite solution, FeCl3 or Na2S. Some of these reducing agents can be toxic to certain anaerobic bacteria. These samples can also be subsampled into plastic Falcon tubes or other plastic containers for immediate freezing at −80°C for nucleic acid extraction.
Enrichment of microorganisms
Anoxic enrichments were set up for isolation of new organisms using a specific basic minimal medium, which had been defined with supplements.
For hypersaline environments, we chose to alter the medium with respect to the high salt concentration. We used the MASE I -adapted ‘HACE’ medium containing following, additional components (per litre): NaCl (130 g), KCl6 (6 g), MgSO4×7H2O (29 g), CaCl2×6H2O (1.1 g), NaBr (0.64 g) and yeast extract (5 g).
The recipe for the proposed standardized MASE media I and II for use in enrichments and culturing is shown in Table 2. MASE I was a very basic medium, which was later augmented with other cations and anions in MASE II.
Table 2. The basal enrichment media used in MASE

a Fe2+ source was added after autoclaving and required a sterile filtration step before adding to the sterile medium. The FeCl2 solution was stored under anoxic conditions.
b The selenite-tungstate solution could be either purchased or prepared according to the following recipe: selenite–tungstate solution per litre: NaOH 0.5 g, Na2WO4×2H2O 4 mg, Na2SeO3×5H2O 3 mg, H2O to 1 litre. Sparge with 100% N2 and filter sterilize.
c Wolfe's Mineral Solution is comprised (Balch et al. Reference Balch, Fox, Magrum, Woese and Wolfe1979): Nitrilotriacetic acid* 1.5 g, MgSO4×7H2O 3.0 g, MnSO4×H2O 0.5 g, NaCl 1.0 g, FeSO4×7H2O 0.1 g, CoCl2×6H2O 0.1 g, CaCl2 0.1 g, ZnSO4×7H2O 0.1 g, CuSO4×5H2O 0.01 g, AlK(SO4)2×12H2O 0.01 g, H3BO3 0.01 g, Na2MoO4×2H2O 0.01 g in 1.0 litre H2O; *Add nitrilotriacetic acid to approximately 500 ml of water and adjust to pH 6.5 with KOH to dissolve the compound. Take volume to 1.0 litre with remaining water and add remaining compounds one at a time. The solution was filter sterilized and stored in dark at 4°C.
d Wolfe's vitamin solution was comprised (Balch et al. Reference Balch, Fox, Magrum, Woese and Wolfe1979): biotin 2.0 mg, folic acid 2.0 mg, pyridoxine hydrochloride 10.0 mg, thiamine HCl 5.0 mg, riboflavin 5.0 mg, nicotinic acid 5.0 mg, calcium D-(+)-pantothenate 5.0 mg, vitamin B12 0.1 mg, p-aminobenzoic acid 5.0 mg, thioctic acid 5.0 mg in 1.0 litre distilled water. The solution is filter sterilized and stored in dark at 4°C.
Mars-relevant supplements
In order to enrich for specialists or specific groups of microorganisms, the medium was supplemented (Table 3). These supplements were added to attempt to enrich a wide range of organisms that use different electron donors and acceptors or had salt tolerance that could be relevant for Mars. The groups for which enrichment was attempted included:
(1) Autotrophs (enriched using C1 compounds, including CO2, CO (carbon monoxide), dimethyl sulphide (DMS), dimethyl amine (DMA), methanol). Enriched on the basis of the presence of these compounds in the Martian atmosphere or the potential endogenous synthesis of simple organics on Mars (e.g. Shock & Schulte Reference Shock and Schulte1998; Steele et al. Reference Steele2012).
(2) Organisms capable of metabolizing amino acids and polycyclic aromatic hydrocarbons (PAHs). Enriched because of the presence of these compounds as components of carbonaceous chondrites (Pizzarello & Cronin Reference Pizzarello and Cronin2000; Sephton Reference Sephton2002; Pizzarello Reference Pizzarello2007) and in the case of PAHs, their detection in Martian meteorites (McKay et al. Reference McKay, Gibson, Thomas-Keprta, Vali, Romanek, Clemett, Chillier, Maechling and Zare1996).
(3) Methanogens (using a hydrogen and CO2 headspace). Enriched on account of the detection of methane on Mars (Webster et al. Reference Webster2015).
(4) Fe-reducers (using oxidized iron compounds). Enriched on account of the presence of iron oxides and other Fe-bearing weathering products on Mars (Bibring et al. Reference Bibring2006).
(5) Fe-oxidizers (using soluble reduced iron). Enriched on account of the presence of olivine and other Fe2+ containing minerals (Hamilton & Christensen Reference Hamilton and Christensen2005).
(6) Sulphate-reducers (using sulphates). Enriched on account of the presence of widespread sulphate minerals on Mars (Vaniman et al. Reference Vaniman, Bish, Chipera, Fialips, Carey and Feldman2004; Gaillard et al. Reference Gaillard, Michalski, Berger, McLennan and Scaillet2013).
(7) Nitrate-reducers (using nitrates). Enriched on account of the presence of fixed nitrogen compounds on Mars (Stern et al. Reference Stern2015).
(8) Fermenters. Capable of degrading biomass from other organisms (using yeast extract as the carbon source in this study). Yeast extract is a mixture of biologically derived molecules that are not expected to be indigenous to Mars in the absence of a biota. However, they would be present in the form of dead biomass if there was a biota.
(9) Salt-tolerant microorganisms. Enriched on account of the orbital detection of evaporitic chloride-deposits (Osterloo et al. Reference Osterloo, Hamilton, Bandfield, Glotch, Baldridge, Christensen, Tornabene and Anderson2008) (Table 3). Halophiles were targeted with HACE medium as described above. Acidophilic microorganisms require low pH values and the medium was adjusted accordingly. In some enrichments, an antibiotic mixture was added to attempt to enrich for archaea by preventing the growth of bacteria.
(10) Acidophiles. Enriched on account of evidence for acidic environments during the history of Mars (e.g. Clark et al. Reference Clark2005).
Table 3. Supplements for MASE I and II and HACE for setup of experiments (percentages in w/v)
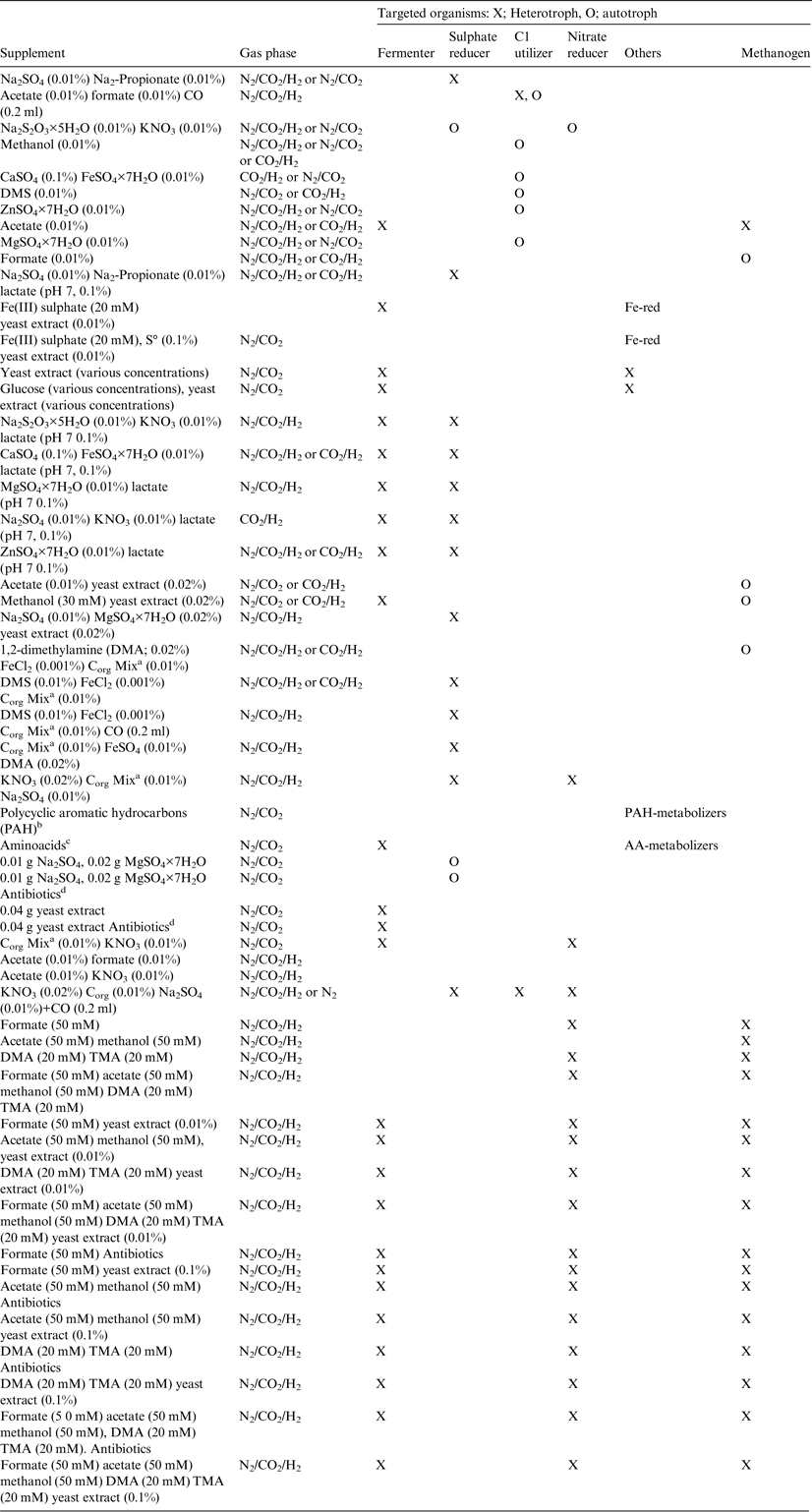
a Corg Mix stock solution: yeast extract 0.5 g, peptone 0.5 g, brain heart infusion 0.5 g, meat extract 0.5 g, 1× Wolfe's Vitamins 500 µl, H2O to 50 ml. The stock solution was filter sterilized.
b The following PAHs and combinations thereof were used per 20 ml: Low Molecular Weight (LMW) PAHs: Acenaphthylene (1 mg); acenaphthene (1 mg); naphthalene (0.1 mg); fluorene (1 mg); anthracene (1 mg); phenanthrene (1 mg); mixture of LMW PAHs (all 1 mg except for naphthalene); mixture of all LMW PAHs (all 1 mg except for naphthalene) with antibiotics. High Molecular Weight (HMW) PAHs: Fluoranthene (1 mg); Pyrene (1 mg); Chrysene (1 mg); Benz[a]anthracene (1 mg); Benzo[a]pyrene (1 mg); mixture of all HMW PAHs; mixture of all HMW PAHs with antibiotics; mixture of all PAHs (1 mg except for naphthalene); mixture of all PAHs (1 mg except for naphthalene) and antibiotics.
c The following aminoacids were added to a final concentration of 10 mM: casaminoacids, N-ethylglycine, D-norvaline, L-norvaline, D-2-aminobutyric acid, L-2-aminobutyric acid, D,L-2-aminobutyric acid, 2-aminoisobutyric acid, DL-3-aminobutyric acid, 4-aminobutyric acid, L-serine, L-methionine, L-threonine, L-isoleucine.
d Antibiotics (100 ml): carbenicillin 0.2 g, streptomycin 0.2 g, rifampicin, 0.4 g, cephalosporin 0.2 g, H2O. Antibiotics stock solution was filter sterilized and 0.1 ml was added to 20 ml media.
Medium preparation
Liquid medium: The following methods were used to prepare pre-reduced anoxic liquid media (Miller & Wolin Reference Miller and Wolin1974). All ingredients as above, except the reducing agent (cysteine–HCl) were dissolved in distilled H2O and dispensed in a glass bottle. The bottle was sealed air-tight with a rubber stopper and a screw cap with aperture. To remove the majority of the oxygen, the medium was bubbled with N2 for 30 min at room temperature. For the complete reduction of the substances in the medium, the reducing agent was dissolved in 1 ml of medium and subsequently transferred into the bottle. The reduction of the medium was observed by the fading of the redox indicator resazurin from blue to pink to colourless after autoclaving. Subsequently, the pH value was tested again in case there was a shift brought on by the reducing agents and if necessary, it was readjusted. The medium was placed into an anaerobic chamber (Coy anaerobic chamber (Coy Laboratory Products, USA) with an atmospheric gas mixture of N2/H2 = 95 : 5 (v/v)) and in the anaerobic chamber 20 ml were dispensed into serum bottles. Each filled bottle was closed with a rubber stopper, removed from the anaerobic chamber and crimped with aluminium seals. At a gassing station, the preliminary gas phase was replaced with the required gas mixture (N2, N2/H2 = 80 : 20 (v/v), CO2/H2 = 80 : 20 (v/v), CO2/N2 = 20 : 80 (v/v)) under sterile conditions by applying a filter to the gas stream to an internal pressure of 1–1.5 bar.
In the specific case of media containing PAHs, these compounds are insoluble in water. Furthermore, PAHs cannot be autoclaved without them degrading. Therefore, PAHs were diluted in a specific solvent (mainly acetone) and aliquots were pipetted into sterile serum bottles. The bottles were kept in a laminar flow hood until the solvent had evaporated. They were then transferred in the anaerobic chamber and autoclaved medium was added. Once sealed, the bottles were gassed under sterile conditions as above and they were stored until required.
Solid medium: The first steps of the preparation are similar to those of anoxic liquid media. All components, including the agar to a final concentration of 1.5%, except the reducing reagents, were dissolved in water in 1 litre glass bottles. The medium was bubbled for 30 min with N2 and was then reduced with the required reagent. After adjusting the pH, the suspension was autoclaved. When the liquid cooled to the required temperature between 50 and 60°C, the plates were poured in the anaerobic chamber, packed in plastic bags and stored at room temperature within the anaerobic chamber.
Isolation of organisms
The down-stream analysis requires strain purification. Pure cultures from liquid medium were obtained by plating the enrichment culture to solid MASE media. Then an anoxic atmosphere was obtained using an anaerobic jar filled with anoxic gas mix as above. Once growth was observed, colonies were picked and purified via three subsequent streak-outs. Often, strains were not able to grow on solid medium. In these instances, purification was done either using optical tweezers technology (described in Huber et al. Reference Huber, Burggraf, Mayer, Barns, Rossnagel and Stetter1995; Fröhlich & König Reference Fröhlich and König2000), or using a dilution series in liquid medium. A dilution series was done by transferring 0.2 ml of enrichment into fresh medium. Then six, 10-fold dilutions were performed and incubated. The highest dilution that showed growth was used for another dilution series. Cell growth was checked via microscopy.
Phylogenetic identification
DNA extraction from pure isolates was achieved using the peqGOLD Bacterial DNA Kit from PeqLab according to the manufacturer's instructions. 16S rRNA gene amplification was performed using the PCR GoTag mastermix. For amplification the 27F (Lane Reference Lane1991) and 1389uR (Marchesi et al. Reference Marchesi, Sato, Weightman, Martin, Fry, Hiom, Dymock and Wade1998) primer sets were used. In this case, amplicons were obtained after 25 cycles of the following programme: initial denaturation 94°C for 2 min, denaturation 94°C for 2 min, annealing 55°C for 1 min, elongation 72°C for 1.5 min and final elongation 72°C for 10 min. Alternatively, primer sets 9bF /1406uR (Lane Reference Lane1991) and 344aF (Casamayor et al. Reference Casamayor, Massana, Benlloch, Øvreås, Díez, Goddard, Gasol, Joint, Rodríguez-Valera and Pedrós-Alió2002)/1406uR were used for amplification. In this case, amplicons were obtained by initial denaturation at 95°C for 2 min, followed by 10 cycles of denaturing at 96°C for 30 s, annealing at 60°C for 30 s and elongation at 72°C for 1 min, followed by another 25 cycles of denaturing at 94°C for 30 s, annealing at 60°C for 30 s and elongation at 72°C for 60 s and a final elongation step at 72°C for 10 min. The QIAquick PCR Purification kit from QIAGEN was used for purification of the amplicons prior to sending the samples for Sanger sequencing. Partial and complete sequences were analysed by the internet platforms NCBI-BLAST (Altschul et al. Reference Altschul, Gish, Miller, Myers and Lipman1990) and EzTaxon (Chun et al. Reference Chun, Lee, Jung, Kim, Kim, Kim and Lim2007).
Long-term conservation of pure cultures
Glycerol cultures: A colony was picked from a plate and transferred into 5 ml of anoxic liquid medium. This culture was incubated overnight. Five hundred microlitres of the freshly grown culture and 500 µl glycerol (87%) were mixed gently under anoxic conditions in a 2 ml Eppendorf tube and frozen at −20°C.
Glass capillaries: The strains were cultivated in their appropriate media. Then 0.95 ml of a freshly grown culture was removed from a vial using a syringe that already contained 50 µl dimethyl sulfoxide (DMSO). DMSO is an anti-freezing agent used to prevent the formation of ice crystals during thawing of the culture. Approximately three quarters of the sterile glass capillaries were filled with the mixture under a laminar flow hood and the ends were closed by melting with a Bunsen burner flame. The prepared glass capillaries were kept on ice until storage at −20°C. Capillaries were prepared in replicates of five per culture. To make sure that the organisms survived this process, a few days after freezing one representative capillary was thawed and inoculated into the appropriate medium.
Deposition into a culture collection
The open/general culture collections accept biological material of applied, biotechnological, educational, taxonomic and general interest such as bacterial strains from which whole genome sequences have been determined. No accession fee has to be paid and bacterial strains sent for deposit should have been used in work already published or prepared for publication. The biological material deposited in this manner will later on be made available to the international scientific community in both academic and industrial institutions.
In 1996, the European Culture collections established a homepage showing information on common access to biological resources (http://www.cabri.org), including quality guidelines concerning the laboratory procedures applied within the deposition process. Further, the European Culture Collections’ Organization (ECCO, http://www.eccosite.org) has defined the core content of a Material Transfer Agreement (MTA) to be used for the supply of samples from the biological material that ECCO holds in its various public collections. This makes biological material available from ECCO collections under the same core conditions.
These rules can be found on the respective web sites, but we review briefly the main points here. The key items within this MTA are quality, safety and security, traceability, access and benefit sharing. These key items are of upmost importance for depositors as well as for all recipients working with the biological material, e.g. in order to ensure that the use of organisms received complies with the general requirements of the Convention on Biological Diversity (CBD), which entered into force at the end of 1993. Article 15 of the CBD emphasizes the sovereign right of individual countries to facilitate access to their genetic resources. As a consequence permission from the provider country on mutually agreed terms or prior informed consent may be required to get access to the samples from which the genetic resources may subsequently be isolated.
The Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization to the CBD entered into force in October 2014. In order to be in compliance with CBD and Nagoya, the culture collection needs a minimal set of data from a depositor, which will be summarized in an accession form. Information required by the depositor comprises the origin of the genetic resource, the legal status of the sample from which the genetic resource was isolated from and some contractual impacts concerning acquisition, transfer and utilization of the bacterial strain.
Data about the identity of the bacterial strain, risk assessment and growth conditions to be applied must also be provided. A strain cannot be accepted for deposition without having these data available. In case that the accession form is complete, a depositor will be asked to send out a subculture of the strain under question together with a signed copy of the accession form. After receipt of the culture the deposition procedure starts by growing subcultures of the strains to confirm purity and to check its growth behaviour. If possible cultures are subsequently freeze dried and aliquots will also be stored in liquid nitrogen, using the glass capillary method (see above). After processing, viability test must be performed to confirm that the strain survived the process (freeze drying, storage in liquid nitrogen). Subsequently, the identity of the strain is checked by partial 16S rRNA gene sequencing (first 800 bp). Depending on the taxon under question, several other methods can be applied in addition for further characterization: Automated ribotyping (DNA fingerprinting based on the analysis of complete rrn operons), Maldi-TOF MS of whole cells (based on mass spectra of proteins), physiological testing or chemotaxonomic analysis (such as whole fatty acid pattern).
Results
Selection of field sites
Six field sites were chosen for the acquisition of anoxic samples (Table 4). Here we provide the basic rationale for site selection with respect to the follow-on sampling. The sample sites were chosen to represent diverse geochemical conditions and periods during Martian history. All of the sample sites are characterized by generally nutrient poor conditions. The rationale for each site and the extreme conditions relevant to Mars is described in Table 4.
Table 4. Field sites chosen for study in the MASE project

All samples were collected using anoxic microbiology procedures. Some of the major physical conditions relevant are shown and comments on the relevance to Mars are listed. All sites were characterized by generally low organic/nutrient availability. Further detail in text.
Much of early Martian history was characterized by water–rock interactions in basaltic environments causing the formation of weathering products such as phyllosilicates (Bibring et al. Reference Bibring2006; Ehlmann et al. Reference Ehlmann, Mustard, Murchie, Bibring, Meunier, Fraeman and Langevin2011). During this early period of Martian history many of the environments were essentially freshwater, with high water–rock ratios. The anoxic environments of Iceland, where sustained bodies of liquid water interact with volcanic bedrock, with few other rock types represented, is a potential analogue for these early Martian environments (Cousins Reference Cousins2015). Throughout Martian history, Icelandic-like anoxic environments with high water–basaltic rock ratio interactions may have occurred during local volcanic or impact melting of glaciers, but they were particularly prevalent during the pre Noachian to Noachian periods (i.e. >~3.8 billion years ago, Ga) when liquid water was relatively common at the surface of the planet. For this work, we chose Lake Grænavatn in the Reykjanesfólkvangur National Park, near Krýsuvík in Iceland, a small spring-fed lake of approximately 360 × 260 m2 in size, with a maximum depth of 45 m. Three sets of samples were collected along the edges of the Lake (LG I, II and III).
The cold sulphidic springs of the environment of Regensburg (Bavaria, Germany), are part of a network of springs that emanate from the subsurface. The domination of the sulphur cycle on Mars (Gaillard et al. Reference Gaillard, Michalski, Berger, McLennan and Scaillet2013) makes aqueous environments containing diverse sulphur species useful potential analogues. Two sulphidic springs are easily accessible and were chosen for sampling (Sippenauer Moor, Islinger Mühlbach). All sulphidic springs are characterized by low organic content, low temperatures, anoxia and sulphur compounds as a possible energy source. At the Islinger Mühlbach site, samples can be collected from approximately 1 m below the surface, where the upwelling water is not yet mixed with atmospheric oxygen and is strictly anoxic. Sippenauer Moor and Islinger Mühlbach are independent and not connected in the deep subsurface. However, they both emanate from Mesozoic karst formations.
The Boulby Mine is a 1.1 km-deep mine in the north-east of the UK which has been excavated into layers of halite, sylvanite and sulphate salts (‘polyhalite’) laid down in the Permian (~250 Ma ago) in the Zechstein inland sea. The presence of evaporite sequences on Mars containing halite and sulphate salts (Vaniman et al. Reference Vaniman, Bish, Chipera, Fialips, Carey and Feldman2004; Osterloo et al. Reference Osterloo, Hamilton, Bandfield, Glotch, Baldridge, Christensen, Tornabene and Anderson2008; Gaillard et al. Reference Gaillard, Michalski, Berger, McLennan and Scaillet2013) makes Boulby a useful Mars analogue. Water from aquifers above and below the evaporite sequences passes through the salt and generates ponds of saturated salt solutions. In the bottom of these pools sediments of salt crystals and detritus (including clays) from the salt collects forming anoxic sediments. These sediments are potential analogues for anoxic sediments within halite evaporative sequences on Mars (Osterloo et al. Reference Osterloo, Hamilton, Bandfield, Glotch, Baldridge, Christensen, Tornabene and Anderson2008). Samples were collected using the Boulby International Subsurface Astrobiology Lab (Cockell et al. Reference Cockell, Payler, Paling and McLuckie2013) within the framework of the MINAR (Mine Analog Research) Program at Boulby (Payler et al. Reference Payler2016). Two sets of samples were collected from two brine pools (‘Billingham Bath’: pools BB-I and BB-II). The brine from BB-II flowed into the brine pool BB-I.
The Río Tinto is an analogue site in the Iberian Pyritic Belt (IPB), Spain. The IPB is one of the largest massive sulphidic deposits on Earth, formed as a hydrothermal deposit during the Paleozoic accretion of the Iberian Peninsula. Río Tinto is an unusual ecosystem due to its size (near 100 km long), constant acidic pH (mean pH 2.3), high concentration of heavy metals and high microbial diversity (Fernandez-Remolar et al. Reference Fernandez-Remolar, Rodriguez, Gomez and Amils2003, Reference Fernandez-Remolar, Morris, Gruener, Amils and Knoll2005; Sánchez-Andrea et al. Reference Sánchez-Andrea, Rodriguez, Amils and Sans2011). One important characteristic of Río Tinto is the high concentration of ferric iron and sulphates, products of the biooxidation of pyrite, the main component of the IPB system. The acidic weathering of the bedrock results in water rich in iron and sulphur. Exploration by the NASA rover Opportunity has revealed sulphate- and hematite-rich sedimentary rocks exposed in craters and other surface features of Meridiani Planum, Mars (Clark et al. Reference Clark2005; Tosca et al. Reference Tosca, McLennan, Clark, Grotzinger, Hurowitz, Knoll, Schröder and Squyres2005).
Permafrost deposits offer the possibility of isolating organisms from liquid water produced in otherwise permanently frozen locations. Mars is known to host permafrost under its surface, particularly at high latitudes (Bandfield Reference Bandfield2007; Bandfield & Feldman Reference Bandfield and Feldman2008). If this permafrost has melted in its past, then liquid water-rich subsurface permafrost environments are plausible analogue environments for habitable locations on Mars. Permafrost samples were collected from two sites: Herschel Island, Canada and the Yedoma in Russia. Samples were collected from silty clay-rich permafrost on Herschel Island (SlpD14 and TSD 14). Herschel is a 15×by 8 km2 island located in the Beufort Sea off the north coast of Canada's Yukon territory. Permafrost on Herschel is composed of perennially frozen ice-rich sediments. The Yedoma is a region of Pleistocene-age permafrost in the north of Russian Siberia and is characterized by its organic-rich nature (about 2% carbon by mass) with an ice content in the range of 50–90%. Yedoma was formed by glaciation over extremely cold and dry steppe-tundra ecosystems. Samples were obtained from a region of continuous Yedoma permafrost (SOB14).
Similar to permafrost environments, the edges and bedrock of glacial environments offer the possibility of collecting samples from perennially frozen soils and ices. Glacier samples were collected from a rock glacier in the Kaunertal, Austria in the frame of the Austrian Space Forum's AMADEE analog program. The Kaunertal valley is 28 km in length and runs southeast from the town of Prutz to the Kaunertal Glacier. Samples were taken at three different sampling locations at the glacier, either in close proximity (SS2 and 3) or several meters apart (SS1 and SS2/SS3). Two samples were soil samples (SS1 and 2) and one sample was taken from a streamlet (directly next to SS2), derived from melted glacier ice.
Collection of anaerobic microbiology samples
Figure 1 shows the range of samples collected from the field sites in which new samples were acquired for MASE. In addition to the samples collected for the isolation methods described in this paper, other samples were collected. These samples are crucial for carrying out the characterization of the total microbial community and determining the geochemical and environmental context of the collected samples. We do not describe the downstream analysis of these samples here as these data will be published in other papers. These samples included: ‘FISH (Fluorescent In-Situ Hybridization) samples’ to quantify organisms and study their morphology and spatial distribution using DNA probes according to standard FISH procedures, ‘DNA extraction samples’ to undertaken environmental rRNA gene and metagenomics analysis to determine the presence of functional genes and the phylogeny of the total anaerobic population. Samples were collected to determine the geochemical context of the solutions and sediments from which organisms were enriched. They were: ‘DOC analysis’ samples for determining dissolved organic carbon, ‘Cations and anions analysis’ samples to determine inorganic geochemistry, and ‘total nitrogen and total carbon’ samples to determine the presence and abundance of N and C available for the microbial communities. Other samples were also collected for projects within MASE, which were: ‘SOLID (Signs Of Life Detector) samples’ to be analysed by the Spanish Centre for Astrobiology using the SOLID instrument to investigate the use of antibody detection methodologies for detecting organisms and their biomarkers, ‘amino acid extraction’ samples to determine the presence of amino acids and the biosignatures of life.

Fig. 1. Schematic representation showing samples collected from each field site using the methodologies described in this paper. Sample names are discussed in the results.
Enrichment of microorganisms
From the samples collected in Fig. 1 microbial enrichments were obtained using the methodologies described in this paper. For these samples the total number of enrichments obtained is shown in Table 5. In total 1131 enrichments were obtained and 118 yielded growth (10.4%). Among these, 131 enrichments were set up to enrich for autotrophs (no organic carbon supplement). Thirty-four of these displayed growth.
Table 5. Enrichments obtained from the sample sites

The number of enrichments was very large, but only a subset of these samples demonstrated high cell densities suitable for enriching organisms with doubling times appropriate for follow-on experiments. A selection of these enrichments using the MASE II medium with their corresponding cell densities are shown in Table 6 to illustrate the types of enrichments that were acquired from each environment.
Table 6. Enrichments selected from those established in Table 5 from which to obtain isolates showing the culture conditions and gas phase used

Isolation of organisms
The 31 isolates (30 bacteria and one archaea) obtained from high cell density enrichments are listed in Table 7 along with DSMZ numbers of those organisms that at the time of submission of this paper had been submitted to DSMZ. The enrichment conditions from which each isolate was obtained are also shown. We isolated a range of organisms that used carbon sources or redox couples of relevance to Mars, including iron oxidizers and reducers and organisms capable of metabolizing carbon sources expected on Mars including amino acids and PAHs. Among these, acidophiles, psychrotolerant/philic and halophilic organisms were also isolated reflecting the link between not just carbon and energy use, but the tolerance to extreme physical and chemical stresses relevant for Martian environments. We obtained six isolates from Lake Grænavatn and one isolate, Yersinia sp. (MASE-SM-9) for which no organic carbon supplement was used. These may either be true autotrophs or using low levels of residual organic carbon in the medium. In Fig. 2, the phylum and genus-level designation of the isolates is also shown.

Fig. 2. Summary of isolates obtained from enrichment cultures.
Table 7. Isolates obtained from the enrichments (Enrichments were at pH 7 and 32°C unless stated otherwise)

Deposition into a culture collection
The project seeks to deposit as many of its isolates as possible in the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH) under the robust deposition requirements described in the ‘Methods’ section. Table 7 provides the DSMZ numbers of isolates that had been deposited within DSMZ at the time of paper submission.
Further use of organisms
Once organisms are deposited in a culture collection they have a range of further uses. The organisms collected in the framework of MASE have been used by the same consortium to study the process of fossilization and thereby assess the survivability and detectability of biosignatures of anaerobic microorganisms during and after natural fossilization and diagenesis using field and spacecraft instrumentation. These efforts have particular importance in improving our knowledge of the factors that influence microbial preservation and thus the detectability of life in ancient terrestrial rocks and the search for such signatures in Martian rocks.
Discussion
One goal of planetary analogue research is to study the physiology, biochemistry and limits of life in environments analogous to those found in extraterrestrial environments and use these data to better assess the habitability of extraterrestrial environments.
The most effective way to accomplish this goal is to devise a systematic approach to the selection of field sites, the collection of samples, the isolation of organisms and their characterization sufficient to deposit them in public culture collections. This is particularly important in the case of anoxic environments from which the isolation of organisms is challenging and particular methods must be consistently applied to collect and enrich anaerobic organisms. As most extraterrestrial environments of interest as locations for life are thought to be anoxic, finding a standardized approach to the isolation of organisms becomes critical.
In this paper, we have described the rationale and approach to a standard approach in the framework of the European Union's Mars Analogues for Space Exploration (MASE) project. We suggest that a similar approach can be taken in any analogue environment for the collection of anoxic samples for environmental, microbial community analysis and/or isolation of organisms.
The success of these methods depended on several key steps: (1) the identification of field sites from which anoxic samples could be reliably collected; (2) the careful definition of an anaerobic microbiology sampling protocol for use in the field that is logistically plausible under a range of field conditions; (3) the use of a defined medium for all enrichments that would allow for isolates to be grown in the same media in future experiments in which comparisons may be made; and (4) a pre-arranged collaboration with an international culture collection so that organisms obtained in the environments can be made publically available.
The isolates we obtained reflect the difficulty of isolating anaerobic organisms from environmental samples generally (Amman et al. Reference Amman, Ludwig and Schleifer1995). We used a wide range of electron donors and acceptors of relevance to Mars to attempt isolation. We obtained isolates capable of iron-oxidation, fermentation and the use of amino acids and PAHs. Some organisms, such as Yersinia spp. were capable of using diverse carbon sources, including amino acids, PAHs and yeast extract and growing anaerobically and are known to be cosmopolitan in the environments (Chen et al. Reference Chen2010).
The use of organics, including amino acids and PAHs as a carbon and energy source in isolation is justified by the expected presence of organics on Mars. A range of organic materials, including amino acids and PAHs are found in carbonaceous chondrites (Pizzarello & Cronin Reference Pizzarello and Cronin2000; Sephton Reference Sephton2002), although the concentration of these compounds at different locations and depths on Mars is not known. One consequence of the use of organics in enrichments is the selection of organisms that may undergo opportunistic growth in laboratory conditions when nutrient conditions are replete, including spore-forming organisms such as Clostridium. Nevertheless, these organisms can be used as model organisms capable of easy, rapid growth and reaching high cell density in the laboratory to carry out fundamental studies of the responses of anaerobic organisms to stressors such as desiccation and radiation.
The approaches used in this work could be modified in a number of ways. For our future studies we required organisms whose doubling times and cell density per ml were compatible with the need to accomplish other tasks, such as stress tests and fossilization studies within the time frame of the project. As the stress tests involve the study of tolerance to salt and other chemical stressors, we also wished to minimize the number of variables involved in the experiments and so organisms were all enriched in the same medium. These restrictions necessarily limit the diversity of organisms that can be obtained. In studies where the intention is not to do cross-comparisons between organisms then the similarity of media is less critical and a greater number of recipes could be used to maximize the diversity of isolated organisms. For example, a greater range of media to target specific metabolic groups could be used such as media for methanogens (e.g. Khelaifia et al. Reference Khelaifia, Raoult and Drancourt2013), iron-reducers (e.g. Hori et al. Reference Hori, Aoyagi, Itoh, Narihiro, Oikawa, Suzuki, Ogata, Friedich, Conrad and Kamagata2015) and sulphate-reducers (e.g. Bade et al. Reference Bade, Manz and Szewzyk2000) in addition to the ones we used. As the culturable population of any community is generally small (Stewart Reference Stewart2012), there exists a trade-off between the restrictions imposed by the experimental requirements and the maximization of the isolated microbial diversity.
Many of the organisms we have collected belong to genera with an influence wider than astrobiology. The isolated Yersinia spp. belong to the same genus as pathogenic organisms, such as Yersinia pestis, the causative agent of the Plague, or the Black Death (Perry & Fetherston Reference Perry and Fetherston1997), a disease still found in localized places on Earth. Yersinia enterocolitica, an organism widespread in soils and animal populations is the causative agent of yersiniosis, a disease with a diversity of clinical symptoms (Bancerz-Kisiel & Szweda Reference Bancerz-Kisiel and Szweda2015). Similarly, the clostridia encompass a large number of organisms important in agricultural and human diseases and conditions (Hatheway Reference Hatheway1990; Wells & Wilkins Reference Wells, Wilkins and Baron1996). Although these associations may discourage the use of these organisms in astrobiology and their provenance in some environments may be a result of human activity, one serendipitous side product of this work is the provision of anaerobic organisms that widen the diversity of organisms in a culture collection that have relevance to genera and families with agricultural and medical importance. For example, expanding our knowledge of their physiology and biochemistry and producing an ever greater number of fully sequenced and annotated genomes from these organisms offer the potential for enhanced knowledge about the capacities of the groups to which they belong and thus even enhanced methods to combat those organisms associated with pathogenic activity. In our case, an annotated genome of our isolated Yersinia intermedia (DSMZ 102845) has been deposited for public use in Genoscope, for instance. Such an effort illustrates powerfully the links between astrobiological research and environmental and medical science.
Acknowledgements
MASE is supported by European Commission's Seventh Framework Programme (FP7/2007–2013) under Grant Agreement no 607297. Collections of samples at the Boulby Mine were made possible through the ICL Ltd., the STFC Underground Science Facility, the Boulby International Subsurface Astrobiology Laboratory and the MINAR (Mine Analogue Research) program. For permafrost samples we thank George Tanski, lead scientist for the Herschel Island campaign and Jens Strauss, responsible for the Yedoma campaign.











