To the Editor—The incidence of infections due to extended-spectrum β-lactamase (ESBL)-producing organisms is increasing in both children and adults.Reference Murray and Peaper 1 This increase is particularly concerning because ESBL-producing organisms are frequently resistant to antibiotics used in empiric sepsis regimens such as ceftriaxone, cefepime, and piperacillin-tazobactam.Reference Chandramohan and Revell 2 Additionally, plasmids carrying genes encoding ESBLs often harbor additional resistance mechanisms reducing the activity of aminoglycosides and fluoroquinolones.Reference Murray and Peaper 1 , Reference Chandramohan and Revell 2 Thus, children with signs and symptoms of severe infections ultimately found to have infections with ESBL-producing organisms may not receive the most appropriate empiric therapy, increasing the likelihood of poor outcomes.Reference Sick, Tschudin-Sutter, Turnbull, Weissman and Tamma 3
Early recognition of ESBL colonization is important because colonization with ESBLs has been associated with subsequent invasive infections.Reference Goodman, Lessler and Cosgrove 4 Children warranting pediatric intensive care unit (PICU) admission are at particularly high risk for serious infections. An understanding of carriers of ESBL-producing organisms may play an important role in guiding appropriate empiric treatment.
We characterized risk factors for ESBL colonization among children admitted to the PICU by conducting a case-control study among patients admitted to The Johns Hopkins Hospital 45-bed tertiary-care PICU in Baltimore, Maryland. Rectal swabs were obtained from all children admitted to the unit between July 2014 and January 2015. Rectal swabs were inoculated into T-soy broth containing a 30-μg ceftriaxone disk and incubated at 37°C. Within 48 hours of inoculation, 100-μL broth samples with visible turbidity were plated on MacConkey agar with a 30-μg ceftriaxone disk and incubated at 37°C overnight. All recovered isolates within the zone of inhibition (inhibition zone, <23 mm) underwent routine identification and antimicrobial susceptibility testing using the BD Phoenix Automated System (Becton Dickinson Diagnostics, Sparks, MD).
Genomic DNA was extracted from isolates with ceftriaxone minimum inhibitory concentrations (MICs) of ≥2 μg/mL using the DNeasy Blood and Tissue Kit (Qiagen, Germantown, MD) and identification of β-lactamase-encoding genes was assessed using the Check-MDR CT103XL kit microarray-based assay (Check-Points, Wageningen, the Netherlands). Multilocus sequence typing (MLST) was used for amplification and sequencing of 8 housekeeping genes for E. coli and 7 housekeeping genes of K. pneumoniae (http://www.pasteur.fr/mlst).
Case patients were defined as children whose admission surveillance culture grew an ESBL-producing organism. Each case patient was matched to 3 control patients using a random number generator. Potential risk factors for colonization were collected on all patients. Data were extracted from all available inpatient and outpatient medical records from facilities within the Johns Hopkins Health System and from medical records of children who received care at institutions within the Epic Care Everywhere Network, a secure exchange that contains patient medical information from a large number of inpatient and outpatient healthcare networks throughout the United States. The Johns Hopkins University School of Medicine Institutional Review Board approved this study with a waiver of informed consent.
Baseline characteristics of cases and controls were compared using a χ2 or Fisher exact test for categorical variables and the Wilcoxon rank-sum test or Student t test for continuous variables. P values ≤.05 were considered significant. All analyses were performed using Stata, version 13 (StataCorp, College Station, TX).
In total, 854 rectal swabs from unique patients were obtained over the study period; 24 children were found to be colonized with ESBLs (2.8%). bla CTX-M genes were identified in all 21 E. coli isolates, in 1 of 2 K. pneumoniae isolates, and in 1 of 1 E. cloacae isolates. A bla SHV-12 gene was identified in the second K. pneumoniae isolate. Of the 24 ESBLs, 17 (71%) contained bla CTX-M-15-like genes. The predominant circulating clonal strain was ESBL-producing E. coli ST131, which was identified in 63% of isolates.
The 24 ESBL-positive case patients were matched to 72 ESBL-negative control patients (Table 1). Within the previous 6 months, case patients were more likely to have had previous ESBL colonization or infection (17% vs 1%; P = .01) or to have been hospitalized in a high-ESBL-burden foreign country (17% vs 1%; P = .01), including China (n=1), India (n=1), Qatar (n=1), and Saudi Arabia (n=2). Case patients were more likely to have received recently chemotherapy (odds ratio [OR], 4.6; 95% confidence interval [CI], 0.9–22.3) or a hematopoietic stem cell transplantation (OR, 10.1; 95% CI, 1.0–102.7). Furthermore, 9 case patients (38%) developed invasive infections with ESBL-producing organisms on a subsequent clinical culture (4 during the hospital admission and 5 within the subsequent 6 months). No control patients developed subsequent ESBL infections.
TABLE 1 Comparison of Children Colonized and Not Colonized With ESBL-Producing Enterobacteriaceae on Admission to a Pediatric Intensive Care Unit
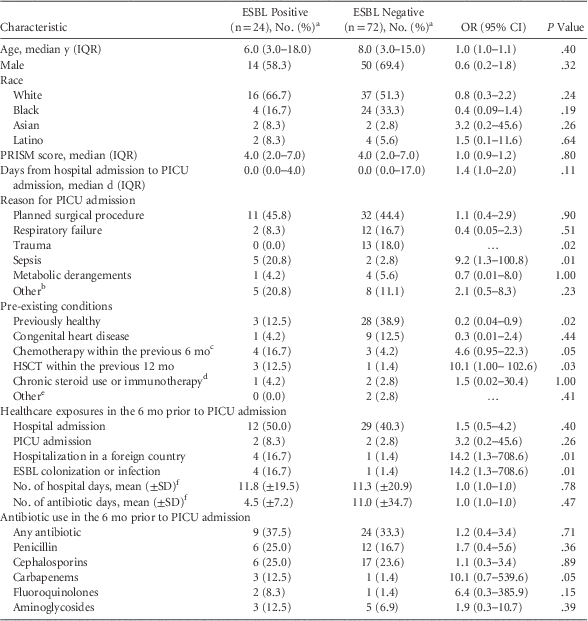
NOTE. ESBL, extended-spectrum β-lactamase–producing; OR, odds ratio; CI, confidence interval; IQR, interquartile range; PRISM, pediatric risk of mortality; PICU, pediatric intensive care unit; SD, standard deviation; HSCT, hematopoietic stem-cell transplantation; SOT, solid organ transplant.
a Unless otherwise noted.
b Other reasons for PICU admission included intractable seizures (n=3), altered mental status (n=2), burns (n=2), myocarditis (n=1), chemotherapy induction (n=1), anemia (n=1), cardiac arrest (n=1), and allergic reactions (n=2).
c Patients who received both chemotherapy and HSCT were only categorized as HSCT.
d Excludes patients receiving immunotherapy for SOT, HSCT, or chemotherapy.
e Other pre-existing conditions include end-stage renal disease on hemodialysis (n=1) and solid organ transplantation (n=1).
f Calculation of the mean included only patients with the specific exposure.
Our findings suggest that targeted screening of high-risk patients may be a reasonable consideration to identify ESBL colonization. Identifying children colonized with ESBL-producing organisms may be particularly relevant to help guide empiric antibiotic therapy because 40% of children colonized with ESBL-producing organisms at the time of PICU admission went on to develop invasive ESBL infections.
ESBL-producing bacteria have increasingly been identified in the community, which appears to be driven by clonal expansion of E. coli ST131 and person-to-person transmission, sometimes in the absence of significant healthcare exposure.Reference Suwantarat, Logan and Carroll 5 – Reference Logan, Meltzer and McAuley 8 Our results indicate that previously healthy children are still at low risk for ESBL colonization. However, hospitalization in a foreign country is a strong predictor of ESBL colonization. This finding reflects that there are significant regional differences in ESBL prevalence, with a disproportionate burden in India, East and Southeast Asia, and the Middle East.Reference Mathai, Rhomberg, Biedenbach and Jones 9 , Reference Tawfik, Alswailem, Shibl and Al-Agamy 10
This study has several limitations. This is a single-center study, and our findings must be repeated in a larger and more diverse setting. Additionally, although we completed a thorough review of inpatient and outpatient records from several healthcare facilities in Maryland, data may have been missing. However, this factor is expected to have been similar for both cases and controls.
As the incidence of ESBLs increases and they contribute to considerable morbidity and mortality, it is imperative to develop systems to identify children who are most at risk of infections caused by ESBL-producing organisms. Our findings suggest that in addition to reviewing prior culture histories to identify children with previous ESBL colonization or infection, targeted screening of children who received medical care abroad in high-risk countries, who recently received chemotherapy, or who recently underwent hematopoietic stem cell transplantations may be another strategy to help identify those most at risk for ESBL colonization.
ACKNOWLEDGMENTS
Financial support: The work was supported by funding the National Institutes of Health (grant no. K23-AI127935) awarded to P.D.T.
Potential conflicts of interest: All authors report no conflicts of interest relevant to this article.



