Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of healthcare-associated infections.Reference Chambers 1 – Reference Magill, Edwards and Bamberg 3 Compared with methicillin-susceptible strains, MRSA is associated with increased risk of adverse health outcomes and increased treatment costs.Reference Cosgrove 4 , Reference de Kraker, Davey and Grundmann 5 Multiple studies have shown that MRSA nasal colonization is a risk factor for subsequent MRSA infection.Reference Huang and Platt 6 – Reference Honda, Krauss and Coopersmith 8 Interventions to interrupt MRSA transmission among hospitalized patients include active surveillance for colonization and contact precautions.Reference Kallen, Mu and Bulens 9 , Reference Burton, Edwards, Horan, Jernigan and Fridkin 10
Recently, the use of chlorhexidine (CHG)-based body wash, either alone or in combination with intranasal antibiotics such as mupirocin, has been demonstrated to reduce the incidence of MRSA transmission and subsequent infection among hospitalized patients.Reference Huang, Septimus and Kleinman 11 – Reference Viray, Morley, Coopersmith, Kollef, Fraser and Warren 13 However, widespread use of chlorhexidine may result in the selection of bacteria that are chlorhexidine tolerant, with the potential to limit the effectiveness of this intervention in the future. In S. aureus, chlorhexidine tolerance is associated with the qac gene family (qacA/B), which code for efflux pumps capable of extruding chlorhexidine and other biocidal compounds from the cell.Reference Mayer, Boos, Beyer, Fluit and Schmitz 14 – Reference Sheng, Wang, Lauderdale, Weng, Chen and Chang 17 The presence of qacA/B has been associated with elevated minimum bactericidal concentrations for chlorhexidine and MRSA decolonization protocol failures.Reference Vali, Davies, Lai, Dave and Amyes 18 , Reference Lee, Macedo-Vinas and Francois 19 The implementation of a daily topical chlorhexidine antiseptic protocol among ICU patients resulted in the selection of a qacA/B–positive (+) MRSA strain, while the transmission of other MRSA strains was reduced during the 2-year period.Reference Batra, Cooper, Whiteley, Patel, Wyncoll and Edgeworth 20 Some plasmids harboring the qac genes may contain additional antibiotic resistance genes.Reference Mayer, Boos, Beyer, Fluit and Schmitz 14 , Reference Sheng, Wang, Lauderdale, Weng, Chen and Chang 17 , Reference Longtin, Seah and Siebert 21 In vitro evidence that CHG may induce selective pressure in clinical MRSA isolatesReference Vali, Davies, Lai, Dave and Amyes 18 , Reference Batra, Cooper, Whiteley, Patel, Wyncoll and Edgeworth 20 highlights the importance of antiseptic tolerance evaluation as part of infection control practices.
Mupirocin, a topical antibiotic, is widely used for treatment of uncomplicated skin and soft-tissue infections and MRSA decolonization. Resistance to mupirocin is broadly classified as low-level resistance (typically defined as having a minimum inhibitory concentration [MIC] of 8–256 mg/L), and high-level resistance (MIC≥512 mg/L)Reference Vali, Davies, Lai, Dave and Amyes 18 , Reference Batra, Cooper, Whiteley, Patel, Wyncoll and Edgeworth 20 , Reference Hetem and Bonten 22 , 23 is most commonly conferred by the plasmid-borne mupA gene, although other mechanisms have been reported.Reference Conly and Johnston 24 – Reference Seah, Alexander and Louie 26 Mupirocin resistance has been associated with reduced effectiveness of decolonization strategies.Reference Walker, Vasquez, Dula and Bullock 27 Its prevalence is variable, ranging from 4% among MRSA isolates in Canadian hospitals to as much as 80% in a Swiss hospital in the context of widespread mupirocin use.Reference Simor, Phillips and McGeer 28 , Reference Jones, Rogers and Brookmeyer 29
Based upon studies of chlorhexidine-based daily bathing protocols and subsequent MRSA infection, this practice is becoming more widespread in hospitals. However, the long-term effects of daily chlorhexidine bathing on the prevalence of qacA/B resistance genes in MRSA isolates in the hospital setting is unknown.
The objective of this study was to determine changes in the frequency of qacA/B gene carriage and mupA-mediated high-level mupirocin resistance among nasal MRSA isolates recovered from active surveillance nasal cultures in the 8-year period during and following implementation of a universal chlorhexidine bathing intervention in a surgical intensive care unit (SICU).
METHODS
Study Design and Setting
We conducted a retrospective cohort study of patients admitted between 2005 and 2012 to the 24-bed surgical intensive care unit of Barnes-Jewish Hospital (BJC), a 1,250-bed tertiary-care hospital in St. Louis, Missouri.
Chlorhexidine Bathing Intervention
On June 1, 2005, the SICU implemented a daily chlorhexidine bathing protocol, replacing a once-daily bath with non-medicated soap and water. Daily water basin baths used 4% chlorhexidine gluconate soap (Exidine, Cardinal Health, McGraw Park, IL). Compliance, as measured by number of soap bottles used per patient day, was >90%. Chlorhexidine soap was diluted in water to achieve a final concentration of approximately 0.125% (1,250 µg/mL). There was no routine use of intranasal mupirocin during this time period.
Data Collection
All patients admitted to the SICU had a swab of the anterior nares obtained for MRSA surveillance cultures per the hospital infection prevention policy. Patients who stayed for >2 days had nasal swabs weekly and at SICU discharge. All MRSA isolates were saved from 2005 to 2012. For this study, isolates from patients admitted to the SICU in 2005 were considered to be collected in the implementation period, and the post-implementation period was 2006–2012.
We estimated a MRSA qacA/B baseline prevalence of 1%, based upon a study of household prevalence in the Saint Louis community.Reference Fritz, Hogan and Camins 30 Thus, we estimated that 63 randomly selected isolates per year (504 total) would be required to detect a 1% per year increase in prevalence of qacA/B positivity during the study period (β=0.80 and α=0.05). Random sampling was done by computer analysis using SPSS version 20 (IBM SPSS, Chicago, IL).
Patient demographics, length of stay in days (LOS, median [interquartile range, IQR]), previous hospitalization history, presence of comorbidities and other characteristics were obtained from the Barnes-Jewish Hospital (BJC) medical informatics database for all patients whose isolates were included in this study. Patients with negative admission cultures but subsequently had a positive nasal MRSA culture during the SICU admission were considered to have ICU-acquired MRSA.
Microbiological Methods
MRSA culture testing was performed as described previously.Reference Warren, Guth, Coopersmith, Merz, Zack and Fraser 31 Isolates were stored in skim milk at −70°C prior to analysis. Selected isolates were subcultured on sheep’s blood agar. MRSA was confirmed using cefoxitin disk diffusion testing according to CLSI standards. 32 Selected samples underwent amplificationReference Mayer, Boos, Beyer, Fluit and Schmitz 14 of the qacA/B genes using a polymerase chain reaction (PCR) with the following primers: F: 5’-CTA TGG CAA TAG GAG ATA TGG TGT -3’, R: 5’- CCA CTA CAG ATT CTT CAG CTA CAT G -3’. As a confirmatory test, a second set of qacA/B PCR was performed on all initially positive isolates.Reference Fritz, Hogan and Camins 30
We performed mupirocin phenotypic susceptibility testing by Kirby-Bauer disk diffusion using a 200-µg mupirocin disk (Oxoid, Hampshire, UK) on Mueller-Hinton agar (Hardy Diagnostics, Santa Maria, CA). 23 Isolates with no zone of inhibition around the mupirocin disk were classified as mupirocin resistant. All mupirocin-resistant isolates underwent PCR amplification for detection of mupA Reference Fritz, Hogan and Camins 30 , 32 , Reference Zhang, Sparling and Chow 33 using the following primers: F: 5’-TAT ATT ATG CGA TGG AAG GTT GG -3’, R: AAT AAA ATC AGC TGG AAA GTG TTG -3’. All MRSA isolates underwent SCCmec typing using a multiplex PCR that could detect and resolve SCCmec types I through V.Reference Boye, Bartels, Andersen, Moller and Westh 34
Statistical Analysis
The Cochran–Armitage trend test was used to compare qacA/B and SCCmec type frequencies by year. Statistical significance was tested using the Mantel-Hansel χ2 test, univariate logistic regression, Student t test, or Wilcoxon signed rank test as appropriate. A 2-sided P≤.05 was considered significant. Statistical analyses were performed with SPSS and SAS v9.3 (SAS Institute, Cary, NC).
This study was approved by the Washington University Human Research Protection Office.
RESULTS
Patient Characteristics
Overall, 1,880 banked MRSA isolates recovered from the anterior nares from 2005 to 2012 were available for testing. In total, 41 isolates were excluded: 27 were repeat isolates from the same patient, 13 were methicillin-susceptible S. aureus on confirmatory testing, and 1 was from a patient who was not in the SICU at the time of testing (Figure 1). After exclusions, MRSA isolates from 504 randomly selected patients (63 per year) were included in the study (Figure 1). Characteristics of the study participants are shown in Table 1. Notably, 70% of the MRSA isolates were obtained within the first 2 days of SICU admission. Among all the MRSA isolates, 36 (7.1%) were qacA/B(+). There was no significant difference in the prevalence of qacA/B MRSA isolates among patients in whom MRSA was initially recovered >2 days after ICU admission (10 [27.8%] qacA/B(+) vs 144 [30.8%] qacA/B(−); P=.85). Also, we found no significant difference in the prevalence of qacA/B positivity among MRSA isolates relative to transfer from another hospital or facility (11 [30.5%] qacA/B(+) vs 91 [19.5%] qacA/B(−); P=.13). Compared with patients colonized with qacA/B(−) MRSA isolates, patients colonized with qacA/B(+) MRSA isolates were less likely to have diabetes (5.6% vs 22.3%; P=.02) or congestive heart failure (0 vs 13.7%; P=.009) (Table 2).

FIGURE 1 Selection of study population. MRSA, methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus; SICU, surgical intensive care unit.
TABLE 1 Patient Characteristics

NOTE. IQR, interquartile range; MRSA, methicillin-resistant S. aureus; CHF, congestive heart failure; COPD, chronic obstructive pulmonary disease; ERSD, end-stage renal disease; HIV, human immunodeficiency virus; BJH, Barnes-Jewish Hospital; ICU, intensive care unit; LOS, length of stay.
TABLE 2 Comparison of Study Participants with MRSA Intranasal Isolates by qacA/B Status

NOTE. IQR, Interquartile range; MRSA, methicillin-resistant S. aureus; CHF, congestive heart failure; COPD, chronic obstructive pulmonary disease; ERSD, end-stage renal disease; HIV, human immunodeficiency virus; BJH, Barnes-Jewish Hospital; ICU, intensive care unit; LOS, length of stay; ER, emergency room.
a Univariate logistic regression (No previous admission as a reference group).
Prevalence of qacA/B(+) and mupA(+)
The prevalence of qacA/B(+) MRSA isolates was 6.2% in the implementation year, then this rate fell to 0–1.5% between 2006 and 2008. The prevalence then increased to 16.9% for 2 years (2009 and 2010) and subsided thereafter, indicating a significant trend for qacA/B (P=.02) (Figure 2). Of the MRSA isolates tested, 35 (6.9%) were mupirocin resistant by the diffusion disk method. All of the phenotypically mupirocin-resistant isolates were mupA(+).

FIGURE 2 Prevalence of qacA/B(+) among sampled MRSA nasal isolates, per year (2005–2012). MRSA, methicillin-resistant S. aureus; p, P value.
The prevalence qacA/B(+) isolates obtained <2 days showed a significant trend compared with isolates obtained ≥2 days of SICU admission (P=.04 for qacA/B <2 days after admission vs P=.29 for qac A/B ≥2 days after admission) (Figure 3).

FIGURE 3 Annual prevalence of qacA/B(+) MRSA, stratified by detection <2 days (n=24) versus ≥2 days (n=12) after ICU admission. MRSA, methicillin-resistant S. aureus; p, P value.
Among all isolates, 9 (1.8%) were both qacA/B(+) and mupA(+). These isolates occurred in 2005 (n=1), 2008 (n=1), 2009 (n=3), 2010 (n=3), and 2012 (n=1). MRSA isolates that were qacA/B(+) were more likely to be mupA(+) than qacA/B(−) isolates (25% vs 5.6%; P=.003) (Table 3).
TABLE 3 Comparison of MRSA Isolate Characteristics by qacA/B Status
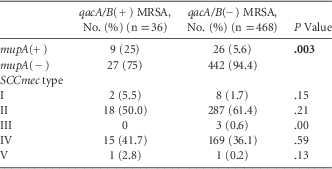
NOTE. MRSA, methicillin-resistant S. aureus; SCCmec, staphylococcal cassette chromosome mec.
Characteristics of qacA/B(+) Isolates Recovered by Year
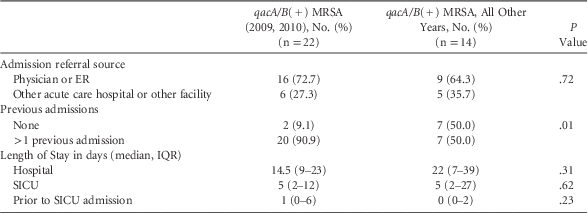
Due to the observed significant trends, we evaluated the data regarding admission source (ie, referred by physician, emergency room (ER), or another facility or acute care facility), hospital LOS, LOS prior to SICU admission, and rate of hospital admissions (previous 12 months) for patients with qacA/B(+) isolates obtained during 2009–2010 and compared them with all other years. We found no significant difference in the admission source (16 [72.7%] vs 9 [64.3%], P=.72 for admission by physician/ER), hospital stay prior to SICU admission (1 [0–6] vs 0 [0–2], P=.23), or total hospital LOS (14.5 [9–23] vs 22 [7–39], P=.11). However, compared with qacA/B(+) isolates from other study years, the qacA/B(+) isolates from 2009 and 2010 were more likely to have ≥1 hospital admissions in the prior year (20 [90.9%] vs 7 [50.0%]; P=.01) (Table 4).
TABLE 4 Comparison of qacA/B(+) Isolates by Year
Prevalence of SCCmec Types
Of the 504 MRSA isolates tested, 10 (2%) were SCCmec type I, 305 (60.5%) were SCCmec type II, 3 (0.6%) were SCCmec type III, 184 (36.5%) were SCCmec type IV, and 2 (0.4%) were SCCmec type V. The prevalence of MRSA isolates that were SCCmec type IV significantly increased from 23.8% in 2005 to 52.4% in 2012 (P<.0001). There was no significant difference in the prevalence of qacA/B(+) by SCCmec type during the study period (Table 3).
DISCUSSION
In our 24-bed SICU, the prevalence of qacA/B genes associated with chlorhexidine tolerance changed in MRSA nasal isolates over the 8-year study period of daily patient bathing with chlorhexidine soap. This change was nonlinear and was associated with an increase in prevalence in years 5 and 6 of the study, then decreased in the remaining 2 years. A comparison of study patients from 2009 and 2010 versus patients from other study years revealed no significant difference in ICU and hospital LOS or rates of hospital admission in the prior 12 months. However, patients from these 2 years were more likely to have a MRSA isolate cultured from a nasal swab collected within the first 2 days of ICU admission (95 of 126 [75.4%] vs 247 of 378 [66.9%]; P=.04). Our comparison of qacA/B(+) isolates from 2009 and 2010 with qacA/B(+) isolates from other study years also revealed no significant difference in admission source, hospital LOS and LOS prior to SICU admission; however, qacA/B(+) isolates from 2009 and 2010 were more likely to have ≥1 hospital admissions in the prior year, suggesting a possibility of an expansion of qacA/B(+) MRSA at BJH during this time period, which resulted in qacA/B(+) patients in those years being exposed in prior admissions. Daily CHG bathing was implemented in 2009 in the other 5 hospital ICUs but not on other inpatient units. There was no marked change in the prevalence of MRSA isolates containing any of the SCCmec type’s genetic element in these 2 years that would suggest a clonal MRSA outbreak in the ICU. We cannot rule out the possibility of dissemination of qacA/B due to horizontal transmission of a qacA/B-containing plasmid among the endemic MRSA strains in the unit.Reference Mathers, Cox and Kitchel 35 However, the prevalence of this genetic element did not remain persistently elevated in the setting of stable daily chlorhexidine use throughout the post-implementation period. Our findings suggest that long-term daily chlorhexidine bathing at the concentration used in this ICU did not result in sustained, widespread dissemination of genes encoding for chlorhexidine tolerance.
The prevalence of high-level phenotypic mupirocin resistance among MRSA nasal isolates was 6.9% and remained stable. This finding is similar to that of an earlier study of nasal MRSA isolates from patients in this ICU from 2002 to 2004, which reported the high-level mupirocin resistance rate to be 8.6%.Reference Jones, Rogers and Brookmeyer 29 Intranasal mupirocin therapy was not systematically used in any area of the hospital during the study period. All high-level mupirocin-resistant MRSA isolates we tested contained mupA. Among the MRSA isolates tested, 9 (1.8%) contained both chlorhexidine and mupirocin resistance determinants. This prevalence is similar to that reported in a study of community-dwelling individuals in St. Louis, Missouri, with a history of skin and soft-tissue infections in which 8 of 755 (1.1%) of MRSA isolates tested contained both resistance elements.Reference Fritz, Hogan and Camins 30 Further study is needed to determine whether the use of universal decolonization regimens using both chlorhexidine and mupirocinReference Huang, Septimus and Kleinman 11 leads to selection for these dually resistant strains in ICU patients.
The SCCmec type IV mobile genetic element has been associated with community-associated MRSA strains in the United States and elsewhere,Reference Deresinski 36 whereas isolates with SCCmec type II have classically been hospital-associated strains. The proportion of MRSA nasal isolates containing the SCCmec type IV mobile genetic element increased steadily over the 8-year study period. This finding is consistent with results from other healthcare facilities,Reference Huang, Tseng, Hu, Tsai, Hsueh and Teng 37 , Reference Nair, Kourbatova and Poole 38 suggesting a shift in epidemiology and that community-associated MRSA strains are now widely disseminated in healthcare facilities.
The present study has limitations inherent to its retrospective design and the use of patient isolates from a single-center site. We were unable to test all the MRSA isolates collected during the study period due to limited resources. However, our random sampling methodology generated a sample with adequate power (β=0.8), which was representative of the patient population admitted during the study period. All of the isolates evaluated in this study were recovered from the anterior nares; thus, we do not know the prevalence of qacA/B in MRSA isolates recovered from other body sites. We did not evaluate phenotypic tolerance to chlorhexidine; therefore, we may have underestimated less common mechanisms of chlorhexidine tolerance.Reference Ye, Zhang, O’Donoghue and Boost 39 , Reference Zmantar, Kouidhi, Miladi and Bakhrouf 40 In addition, we do not know the MIC or minimal bactericidal concentration (MBC) of the strains in this study possessing qacA/B. Chlorhexidine tolerance among Gram-negative bacteria was not examined in our study. Our study was performed in a single ICU; thus, generalizability of results to other facilities may be limited.
Our study has several strengths, including a long follow-up period to evaluate the prevalence rate, the use of both phenotypic and genotypic methods to assess mupirocin resistance, a standardized surveillance protocol for collection of MRSA nasal isolates, and a well-established chlorhexidine bathing program in the ICU.
In conclusion, prevalence of qacA/B associated with chlorhexidine tolerance did change over time among colonizing MRSA isolates over the 8-year period of daily patient bathing with chlorhexidine soap; however, this was not a linear relationship. This change in the frequency of qacA/B genes is most likely due to patients in those years being exposed in prior admissions. The prevalence of MRSA with SCCmec type IV increased over the study period. Further studies are needed to determine whether other ICU-based decolonization strategies, such as universal treatment with chlorhexidine and intranasal mupirocin, will result in selection of co-resistant isolates.
ACKNOWLEDGMENTS
We thank Dr. Margaret Olsen for her valuable advice on study design and the Barnes-Jewish Hospital Clinical Microbiology laboratory for ongoing efforts to support infection prevention and surveillance programs at Barnes-Jewish Hospital.
Financial support: This work was supported by the Prevention Epicenter Program from the Center for Disease Control and Prevention (1U54CK000162-01).
Potential conflicts of interest: D.K.W. has served a consultant to Centene Corporation, Sagentia, and Novaerus Corporation. C.A.B. has served a consultant to ThermoFisher. J.E.M. has served a consultant Astra-Zeneca, Cubist, Pfizer, Merck and Bayer, and has received grant support from Astra-Zeneca, Merck and Bayer. All other authors report no conflicts of interest relevant to this article.









