To the Editor—Elizabethkingia anophelis is a rapidly emerging nosocomial pathogen reported to cause bacteremia in immune-compromised elderly people and neonates.Reference Lau, Wu and Teng 1 , Reference Lau, Chow and Foo 2 The unknown pathogenesis and unclear resistance mechanism of E. anophelis and their phenotypic similarity to E. meningoseptica mislead and complicate the infection management of this pathogen, resulting in treatment failure. Inherent resistance to multiple classes of drugs and absence of an antibiotic sensitivity profile standard for this bacterium makes empirical treatment nearly impossible. Elizabethkingia anophelis bacteremia has recently been considered clinically significant, leading to high morbidity and mortality that has been mistakenly attributed to E. meningoseptica because of their phenotypic similarity.Reference Lau, Chow and Foo 2 Molecular epidemiological analyses of recent Elizabethkingia bacteremia infections and outbreaks have been conducted in United States, Singapore, China, and Korea. These outbreaks were predominated by E. anopheles. Reference Han, Kim and Lee 3 – Reference Wang, Gao and Lin 6 This finding warrants the implementation of molecular typing for an accurate diagnosis to guide appropriate antibiotic regimen instead of relying solely on conventional phenotypic identification with a compact automated VITEK-2 system, which uses a factory default database and lacks timely amendments.Reference Han, Kim and Lee 3 , Reference Chew, Cheng, Lin and Teo 5
In the first report of an outbreak in a tertiary healthcare center of Eastern India, the clinical and molecular epidemiology of 9 bacteremia episodes during 2 months of surveillance from August to September 2017 were identified as E. meningoseptica by the VITEK-2 system. These findings were genetically validated by species-specific markers, such as lipid-A disaccharide synthase gene for E. anophelis and sodium-proton antiporter for E. meningoseptica,Reference Chew, Cheng, Lin and Teo 5 and 16s rRNA gene sequencing. An antibiotic susceptibility study was conducted using the VITEK-2 compact automated system (BioMerieux) with the GN-AST-N280 card. Sensitivity was interpreted according to Clinical Laboratory Standards Institute (CLSI) guidelines (2013). 7 The clonal relatedness among 9 isolates was investigated using repetitive-element polymerase chain reaction (rep-PCR) and (GTG)5 PCR according to the method described by Adiguzel et al.Reference Adiguzel, Ozkan, Baris, Inan, Gulluce and Sahin 8
Nonrepeated Elizabethkingia spp (EA1-9) were isolated from 9 inpatients, and we analyzed the demographic data, clinical characteristics, and outcomes for these cases (Table S1 online). In these 9 cases, Elizabethkingia bacteremia prevailed mostly among elderly people (n=8; median age 52 years), but 1 patient was a 2-year-old child. The male: female ratio among these patients was 7:2 (Table S1 online). Overall, 5 Elizabethkingia isolates were obtained from blood; the rest were obtained from tracheal aspiration (n = 3) and cerebrospinal fluid (n = 1). All of these patients were reported as having hospital-acquired, clinically significant bacteremia, with a high mortality rate (33.3%). Of these 9 patients, 3 died within 1 month of their hospital stay despite treatment with antibiotics (eg, quinolones, penicillin, cephalosporins, carbapenems, etc, either alone or in combination) due to several associated complications: pneumonia, lower respiratory infection, meningitis, acute kidney injury, and metabolic encephalopathy, etc.
These isolates showed resistance to different groups of antibiotics with varying percentages ranging from ~80% to 100% (Table S2 online). However, the highest susceptibility was found against tigecycline and piperacillin-tazobactam, which corroborates the previous reportsReference Han, Kim and Lee 3 , Reference Perrin, Larsonneur and Nicholson 4 , Reference Lin, Lai, Yang, Huang and Lin 9 except isolate EA1, which matched a single study from China.Reference Wang, Gao and Lin 6 The resistance profile against levofloxacin was analogous to isolates identified in Korea and Wisconsin.Reference Han, Kim and Lee 3 , Reference Perrin, Larsonneur and Nicholson 4 However, the alteration of the antibiotic resistance profile depends generally on different types of stress on different sources of Elizabethkingia isolates.Reference Wang, Gao and Lin 6
All 9 isolates were identified as E. meningoseptica by the VITEK-2 compact automated system. Because the identification of Elizabethkingia spp has been reported to be misleading using the VITEK 2 and MALDI-TOF MS systems,Reference Lau, Chow and Foo 2 these samples were subjected to genotypic validation. However, upgrading the VITEK-2 system with better antibiotic sensitivity profiles, updating the CLSI guidelines, and expanding the database for MALDI-TOF mass spectra of E. anophelis will improve their proper identification. All 9 Elizabethkingia spp showed amplification of lipid-A disaccharide synthase gene, a species-specific primer of E. anopheles, and were further confirmed to be E. anophelis by 16s rRNA gene sequence analysis (GenBank accession no: MH121154-MH121158, MN038050-MN0380053). Rep PCR- and (GTG)5 PCR–based phylogenetic analysis of 9 isolates revealed a close clustering of EA1 and EA2 with EA4; EA6 with EA8; and EA5 with EA7. These findings explain the considerable clonal similarity among E. anophelis isolates belonging to the same in-patient departments (Fig. 1).
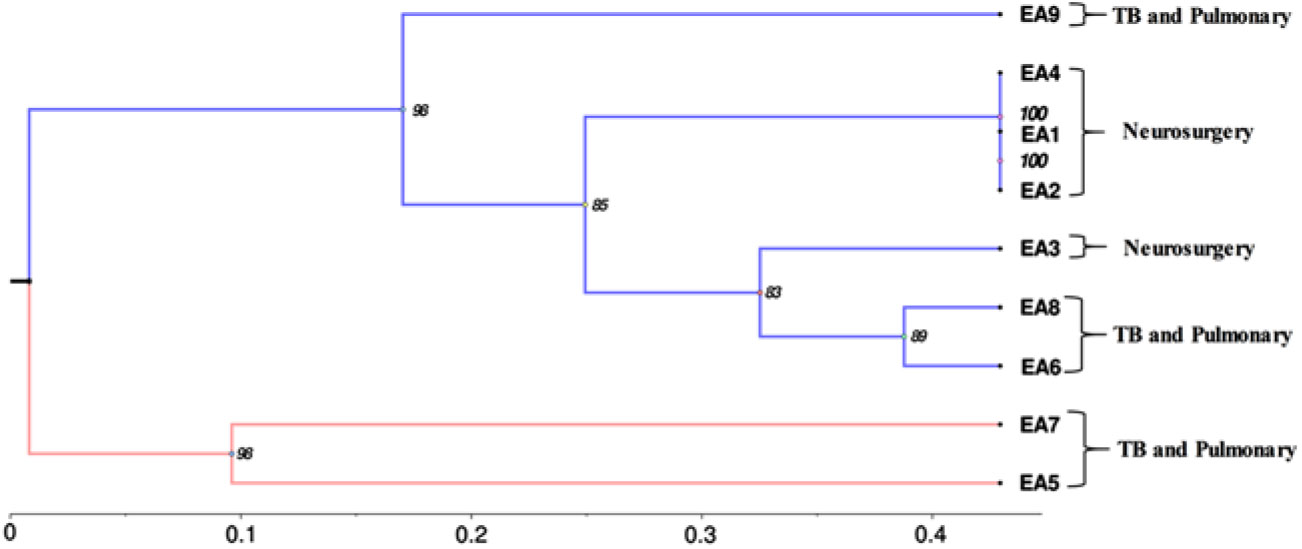
Fig. 1. The phylogenetic tree was developed using REP and (GTG)5 PCR. We observed close clustering among 9 isolates as they relate to the same in-patient department (as labeled): EA1 and EA2 with EA4; EA6 with EA8; and EA5 with EA7. A phylogenetic tree was constructed based on Jaccard similarity coefficient. Hierarchical clustering was performed by hclust function (R Core Team stats 3.5.3) using the unweighted pair group method with arithmetic mean (UPGMA) method, and the tree was cut into an optimal number of clusters along with bootstrapping 1,000 times (Pvclust version 2.0).
This study is the first molecular epidemiological report on a bacteremia outbreak of India with prevalence of E. anophelis bacteria establishing E. meningoseptica to be the more remote cause of bacteremia infection. However, future prospective studies with population-based data over longer surveillance periods should be performed to determine the prevalence and incidence of E. anophelis bacteremia. A repeated molecular epidemiological study should be employed for accurate diagnosis and appropriate treatment regimen.
Acknowledgment
The authors thank Dr N. K. Debata, Department of Microbiology, IMS and SUM Hospital, Siksha ‘O’ Anusandhan University, Bhubaneswar, Odisha, India for providing bacterial samples and related information. The authors are also grateful to Prof (Dr) S. C. Si, Dean, Centre for Biotechnology and Prof (Dr) M. R. Nayak, President, Siksha ‘O’ Anusandhan University, for providing infrastructure and encouragement throughout the study.
Financial support
This study was supported by the Science and Engineering Research Board (grant no. EMR/2016/006732), New Delhi, India.
Conflicts of interest
The authors declare that they have no conflict of interest.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2019.213



