1. Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide, stemming in part from the lack of effective early detection schemes impacting survival (Kadara et al., Reference Kadara, Kabbout and Wistuba2011). There is an urgent need for novel biomarkers that could be clinically applied as prognostic factors and somatic mutations are obvious candidates for being used as prognostic markers (Li et al., Reference Li, Kung, Mack and Gandara2013; Travis et al., Reference Travis, Brambilla and Riely2013).
Epidermal growth factor receptor (EGFR) plays an important role in cell proliferation and survival (Reungwetwattana et al., Reference Reungwetwattana, Weroha and Molina2012). The frequency of activating mutations of the EGFR gene in non-small cell lung cancer (NSCLC) varies according to ethnicity and is noted in 15% of NSCLC diagnosed in Caucasians, 40% in Asians and 33·3% in Latin Americans mostly of Spanish origin (Paez et al., Reference Paez, Jänne, Lee, Tracy, Greulich, Gabriel, Herman, Kaye, Lindeman, Boggon, Naoki, Sasaki, Fujii, Eck, Sellers, Johnson and Meyerson2004; Leidner et al., Reference Leidner, Fu, Clifford, Hamdan, Jin, Eisenberg, Boggon, Skokan, Franklin, Cappuzzo, Hirsch, Varella-Garcia and Halmos2009; Rosell et al., Reference Rosell, Moran, Queralt, Porta, Cardenal, Camps, Majem, Lopez-Vivanco, Isla, Provencio, Insa, Massuti, Gonzalez-Larriba, Paz-Ares, Bover, Garcia-Campelo, Moreno, Catot, Rolfo, Reguart, Palmero, Sánchez, Bastus, Mayo, Bertran-Alamillo, Molina, Sanchez and Taron2009; Arrieta et al., Reference Arrieta, Cardona, Bramuglia, Gallo, Campos-Parra, Serrano, Castro, Avilés, Amorin, Kirchuk, Cuello, Borbolla, Riemersma, Becerra and Rosell2011; Cote et al., Reference Cote, Haddad and Edwards2011).
Activating KRAS mutations that predominantly cluster to either codons 12 and 13, and rarely in codon 61 (Suda et al., Reference Suda, Tomizawa and Mitsudomi2010) are encountered with differing rates in NSCLC diagnosed in different ethnic groups: 30% in Caucasians, 10% in East Asians and 16·6% in Latin Americans (Hunt et al., Reference Hunt, Strimas, Eyer, Haddican, Luckett, Ruiz, Axelrad, Backes and Fontham2002; Riely et al., Reference Riely, Kris, Rosenbaum, Marks, Li, Chitale, Nafa, Riedel, Hsu, Pao, Miller and Ladanyi2008; Arrieta et al., Reference Arrieta, Cardona, Bramuglia, Gallo, Campos-Parra, Serrano, Castro, Avilés, Amorin, Kirchuk, Cuello, Borbolla, Riemersma, Becerra and Rosell2011). Mutations in KRAS and EGFR that appear to be mutually exclusive are currently being used as molecular biomarkers for determining both prognosis and therapeutic targets in NSCLC (Murray et al., Reference Murray, Timotheadou, Linardou, Vrettou, Kostopoulos, Skrickova, Papakostantinou, Christodoulou, Pectasides, Samantas, Papakostas, Skarlos, Kosmidis and Fountzilas2006; Irmer et al., Reference Irmer, Funk and Blaukat2007; Mok et al., Reference Mok, Wu, Thongprasert, Yang, Chu, Saij, Sunpaweravong, Han, Margono, Ichinose, Nishiwaki, Ohe, Yang, Chewaskulyong, Jiang, Duffield, Watkins, Armour and Fukuoka2009; Brevet et al., Reference Brevet, Johnson, Azzoli and Ladanyi2011; Heigener & Reck, Reference Heigener and Reck2011). Tyrosine kinase inhibitors (TKIs) are widely used as an adjuvant treatment to chemotherapy in advanced stage NSCLC cases that specifically display activating EGFR mutations (Keedy et al., Reference Keedy, Temin, Somerfield, Beasley, Johnson, McShane, Milton, Strawn, Wakelee and Giaccone2011). NSCLC harbouring activating KRAS mutations, are resistant to EGFR-TKIs treatment and patients have shorter survival and response rates (Pao et al., Reference Pao, Wang, Riely, Miller, Pan, Ladanyi, Zakowski, Heelan, Kris and Varmus2005; Borràs et al., Reference Borràs, Jurado, Hernan, Gamundi, Dias, Martí, Mañé, Arcusa, Agúndez, Blanca and Carballo2011; Gaughan & Costa, Reference Gaughan and Costa2011).
BRAF is mutated in a wide range of human cancers, including lung cancers (Xing, Reference Xing2005; Dhomen & Marais, Reference Dhomen and Marais2009; Gaughan & Costa, Reference Gaughan and Costa2011). Ninety per cent of BRAF mutations are represented by a single somatic mutation BRAF V600E (Cantwell-Dorris, Reference Cantwell-Dorris2011; Gaughan & Costa, Reference Gaughan and Costa2011) and approximately 50% of lung adenocarcinomas harbour the recurrent somatic oncogenic mutations in EGFR, KRAS and BRAF (Girard, Reference Girard2013; Oxnard et al., Reference Oxnard, Binder and Jänne2013).
Mutations in other genes are involved in modulating therapeutic response to inhibitors of the EGFR/PI3K/AKT pathway (Su et al., Reference Su, Dias-Santagata, Duke, Hutchinson, Lin, Borger, Chung, Massion, Vnencak-Jones, Iafrate and Pao2011). PTEN is a dual specificity phosphatase which directly antagonizes the phosphatidylinositol-3 kinase (PI3K) signalling pathway (Endersby & Baker, Reference Endersby and Baker2008; Tang et al., Reference Tang, Iijima, Kamimura, Chen, Long and Devreotes2011). PTEN is a tumour suppressor gene that is frequently mutated in human cancer, with most mutations leading to an inactivation of the gene (Cantley & Neel, Reference Cantley and Neel1999; Simpson & Parsons, Reference Simpson and Parsons2001; Pandolfi, Reference Pandolfi2008). Although inactivating PTEN mutations are present in approximately 10% in NSCLC, PTEN expression is diminished in a larger proportion of these tumours (almost 70%) possibly by epigenetic mechanisms (Tang et al., Reference Tang, He, Guo and Chang2006; Li et al., Reference Li, Zhao, Peng, Liang, Deng and Chen2012).
TTF-1 is a DNA-binding protein found to be expressed in lung cells, regulates the activity of proliferating cells and also plays a role in angiogenesis (Berghmans et al., Reference Berghmans, Mascauxa, Hallerb, Meerta, Van Houttec and Sculier2008; Wislez et al., Reference Wislez, Antoine, Baudrin, Poulot, Neuville, Pradere, Longchampt, Isaac-Sibille, Lebitasy and Cadranel2010). TTF-1 was reportedly over-expressed in approximately 80% of lung adenocarcinomas (Maeshima et al., Reference Maeshima, Omatsu, Tsuta, Asamura and Matsuno2008). An activating EGFR mutation is associated with TTF-1 over-expression in lung adenocarcinoma (Yatabe et al., Reference Yatabe, Kosaka, Takahashi and Mitsudomi2005; Tapia et al., Reference Tapia, Savic, Bihl, Rufle, Zlobec, Terracciano and Bubendorf2009) and the pattern of EGFR(+)/TTF-1(–) seems to be an exclusive signature for NSCLC, especially squamous-cell carcinoma (SCC) subtype (Berghmans et al., Reference Berghmans, Mascauxa, Hallerb, Meerta, Van Houttec and Sculier2008).
Ethnicity has been shown to affect risk for developing lung cancer. African Americans have higher incidence rates for lung cancer and family history of lung cancer, compared with pack/year smokers matched Caucasians (Cote et al., Reference Cote, Kardia, Wenzlaff, Ruckdeschel and Schwartz2005; Haiman et al., Reference Haiman, Stram, Wilkens, Pike, Kolonel, Henderson and Le Marchand2006). Moreover, for lung cancer patients, being of Japanese ethnicity and never-smoker are independent favourable prognostic factors for overall survival compared with Caucasians (Kawaguchi et al., Reference Kawaguchi, Matsumura, Fukai, Tamura, Saito, Zell, Maruyama, Ziogas, Kawahara and Ignatius2010). These ethnic differences are in all likelihood the result of the combined differences in the rate of germline and somatic sequence alterations in genes involved in NSCLC pathogenesis.
The spectrum of somatic EGFR, KRAS, BRAF, PTEN mutations and TTF-1 expression and its potential associations in genetically heterogeneous Brazilian lung cancer patients has not been previously reported, and that was the focus of this study.
2. Materials and methods
(i) Subjects
The study cohort encompassed 88 patients diagnosed with NSCLC who were eligible for surgery, with no previous history of chemotherapy or radiotherapy. Patients were recruited from a referral centre of thoracic surgery (Hospital Julia Kubitscheck – FHEMIG, Belo Horizonte, Brazil) between 1 January 2006 and 31 December 2011. Controls were 28 healthy individuals older than 55 years with no previous personal or family history of cancer, randomly recruited from the outpatient clinic in the same medical centre during the same time period. A group of 96 healthy Brazilian individuals, representative of Southeastern Brazil, were used as controls for genomic ancestry.
(ii) Ethics statement
The Ethics Committee of Universidade Federal de Minas Gerais (Comitê de Ética em Pesquisa da UFMG, # 373-05) approved the study protocol and all participants signed a written informed consent.
(iii) EGFR/KRAS/BRAF/PTEN genotyping
Genomic DNA of participating NSCLC patients was isolated from fresh tumour tissue samples as well as from peripheral blood, according to a proteinase K-based standard protocol (Miller et al., Reference Miller, Dykes and Polesky1988). Peripheral blood was collected in vacuum tubes and genomic DNA was isolated using the high salt method of Lahiri and Nurnberger (Lahiri & Nurnberger, Reference Lahiri and Nurnberger1991) and was extracted from all study participants – NSCLC cases and controls. Genotyping for germline and somatic alterations was carried out for the following genes and mutations: exons 18 (G719S), 19 (746_750del, D761Y and L747S), 20 (insertions and T790M) and 21 (L858R and L861Q) of EGFR, exons 2 and 3 of KRAS (codons 12, 13 and 61), exon 15 of BRAF (BRAFV600E) and all nine exons of PTEN (to evaluate any inactivating mutation in the entire coding regions of the gene) were amplified by PCR with primers specific for each region (Table 1). For PCR reactions 2 μl of DNA at 30 ng/μl were mixed with 2·5 μl of 10X IIB buffer (40 mm NaCl; 10 mm Tris–HCl pH 8·4; 0·1% Triton X-100; 1·5 mm MgCl2), 2·5 μl of 0·2 mm dNTP, 0·5 μl of each primer at 10 pmol/μl and 0·25 μl of Taq polymerase (Invitrogen, Brazil) 0·625 U, on a final volume of 25 μl. Samples were placed on an Eppendorf Mastercycler® (Hamburg, Germany) at 94°C for 3 min and then 35 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s and a final extension time at 72°C for 5 min. PCR products were purified using Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare, São Paulo, Brazil) following manufacturer's protocol and visualized on a silver-stained 6·5% polyacrylamide gel. To improve the sensitivity of the KRAS mutation detection we used the COLD-PCR method as previously described (Zuo et al., Reference Zuo, Chen, Chandra, Galbincea, Soape, Doan, Barkoh, Koeppen, Medeiros and Luthra2009).
Table 1. Characteristics of PCR amplification of EGFR and KRAS

a Tm: annealing temperature.
Sequences were obtained on ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA). Bidirectional sequence data were analysed using Sequencer 4.9 software (http://genecodes.com). Positive findings for EGFR exon 19 deletions were also confirmed by fragment analysis. The genomic fragment including all exon 19 was amplified in 10 μl final PCR volume of the following: 1X PCR buffer (10 mm Tris–HCl pH 8·3 or pH 9·2, 75 mm KCl, 3·5 mm MgCl2), 200 μ m dNTPs, 1·0 U of Platinum Taq DNA polymerase (Life Technologies, São Paulo, Brazil), 20 ng of genomic DNA, 1·5 μ m of M13-40 forward primer labelled with the FAM dye, 1·5 μ m of each unlabelled reverse primer and 0·1 μ m of each unlabelled forward primer.
(iv) Immunohistochemistry of TTF-1
Tissue sections from 27 samples previously diagnosed by pathological report as SCCs, three adenocarcinomas and four diagnosed as NSCLC poorly differentiated carcinoma were stained with TTF-1 antiserum. Briefly, 4 μm paraffin-embedded sections were dewaxed in xylene and hydrated with graded ethanol. Endogenous peroxidase activity was blocked with 3% H2O2 in water for 10 min. Heat-induced epitope retrieval was performed with 1 mm EDTA buffer pH 8·0 for 30 min in a steamer at 96°C. Primary polyclonal rabbit antiserum was used at 1 : 100 for 18 h at 4°C. This was followed by incubation with the labelled streptavidin-biotin kit NovoLinkTM Max Polymer (Novocastra, UK). Peroxidase activity was developed with DAB (Sigma, St Louis, MO) with timed monitoring using a positive control sample. The sections were then counterstained with haematoxylin, dehydrated and mounted. All slides were examined under light microscopy and staining for TTF-1 was evaluated according to the presence or not of the protein by two pathologists who were blinded to the clinical course of the patient.
(v) Genomic ancestry analysis
Germline DNA of all 88 lung cancer patients and 96 ethnically diverse Brazilian controls were genotyped with a set of 40 biallelic short insertion/deletion polymorphisms (In/Dels), as previously described (Bastos-Rodrigues et al., Reference Bastos-Rodrigues, Pimenta and Pena2006). Amplicons were size fractionated using an ABI 3130 DNA sequencer (Applied *Biosystems) and analysed using the GeneMapper® Software (version 3.7). To estimate the proportion of European, African and Amerindian biogeographical ancestry of each individual we used the Structure program, version 2.3 (http://pritch.bsd.uchicago.edu/structure.html).
(vi) Statistical analysis
The proportion of European, African and Amerindian bio-geographical ancestry of each individual was used for stratifying statistical analysis. For statistical comparisons between cases and controls, the two-tailed Mann–Whitney U test was used. Single-marker allelic and genotypic association tests were performed using the Unphased software package version 3.0.12 (www.mrcbsu.cam.ac.uk/personal/frank/software/unphased/). A value of P ⩽ 0·05 was considered statistically significant. Odds ratios and 95% confidence interval were calculated.
3. Results
(i) Sample characteristics
Demographics and relevant clinical and pathological data of all 88 NSCLC cases are shown in Table 2. The control group (for EGFR germline mutation genotyping) encompassed 28 healthy individuals, comprising 64% women and 36% men with a mean age of 72·8 ± 9·15 years (range 55–89 years).
Table 2. Clinical data of non-small cell lung cancer (NSCLC) patients
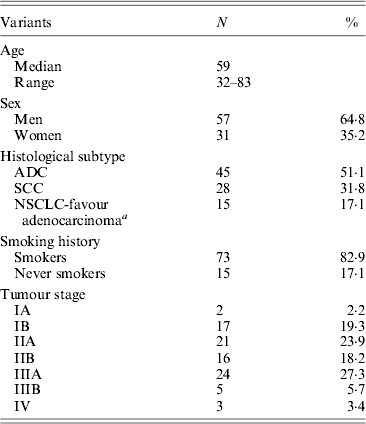
a Tumours positive for TTF-1 marker (Travis et al., Reference Travis, Brambilla and Riely2013).
ADC, adenocarcinoma; SCC, squamous-cell carcinoma.
(ii) EGFR/KRAS/BRAF/PTEN mutation status
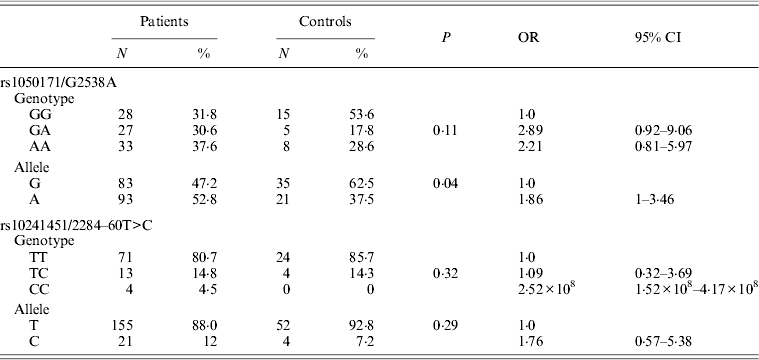
EGFR gene genotyping showed the presence of the 746_750del (LREA domain) in 3/88 patients (3·4%), and was exclusively detected in 3/45 (6·6%) of patients with adenocarcinoma. No sequence alterations, especially G719S, were noted in exon 18 of the EGFR gene. Only one patient diagnosed with adenocarcinoma (1·5%) showed the L585R variation in exon 21 of EGFR. In addition to these clearly pathogenic mutations in the EGFR gene, two previously reported polymorphisms were also identified in DNA of cases and controls: a silent base substitution (CAG>CAA) at c.2538 position (corresponding to rs1050171) was detected in 61·3% of the cases (n = 54) and a substitution at IVS-60T>C position (rs10241451) was noted in 20·4% of the cases (n = 18). The rs10241451 did not show a significant association with the disease both by allele and genotype frequency (P = 0·29 and P = 0·32, respectively) when compared with controls, whereas a significant association between rs1050171 and lung cancer compared with cancer-free controls was noted only by allele and not by genotype frequency (P = 0·04 and P = 0·11, respectively) (Table 3).
Table 3. Allele and genotype frequencies of rs1050171 and rs10241451 in patients and controls
Codon 12 KRAS mutations were noted in five male patients (5·7%) diagnosed with adenocarcinoma: three harboured the Gly12Cys (c.34G>T) mutation and two the Gly12Asp (c.35G>A) mutation. No mutations in codon 13 of KRAS were found. Two female smokers patients (3%) diagnosed with SCC at ages 60 and 63 years old showed a previously reported missense mutation (rs17851045) in codon 61 (c.182A>T), which leads to histidine for glutamine change (H61Q). No sequence alterations in exons 1–9 of the PTEN gene were noted and the BRAF V600E mutation was not detected in any of the samples analysed.
(iii) TTF-1 immunohistochemistry
All three adenocarcinoma samples, which served as positive controls for TTF-1 expression, demonstrated positivity, as expected. Eleven of the 27 SCCs analysed displayed positive TTF-1 expression and in 16 tumours no expression was present (Table 2). Four tumours previously described as poorly differentiated carcinoma also showed positive TTF-1 expression. All positive tumours for TTF-1 expression (n = 15) were considered NSCLC-favour adenocarcinoma.
(iv) Genomic ancestry analysis
We genotyped germline DNA from all 88 lung cancer patient samples and 96 controls, for 40 polymorphic In/Del loci which form a powerful ancestry informative test battery (Bastos-Rodrigues et al., Reference Bastos-Rodrigues, Pimenta and Pena2006). For the case group, the proportions of European, African and Amerindian ancestry were 0·87 ± 0·02 (mean±se), 0·09 ± 0·02 and 0·04 ± 0·007, respectively, whereas for the control group, the results were 0·85 ± 0·02, 0·08 ± 0·01 and 0·07 ± 0·001, respectively. The proportion of European, African and Amerindian ancestry in each group indicated that an African component was more prevalent in lung cancer patients than controls (P = 0·03) (Fig. 1).
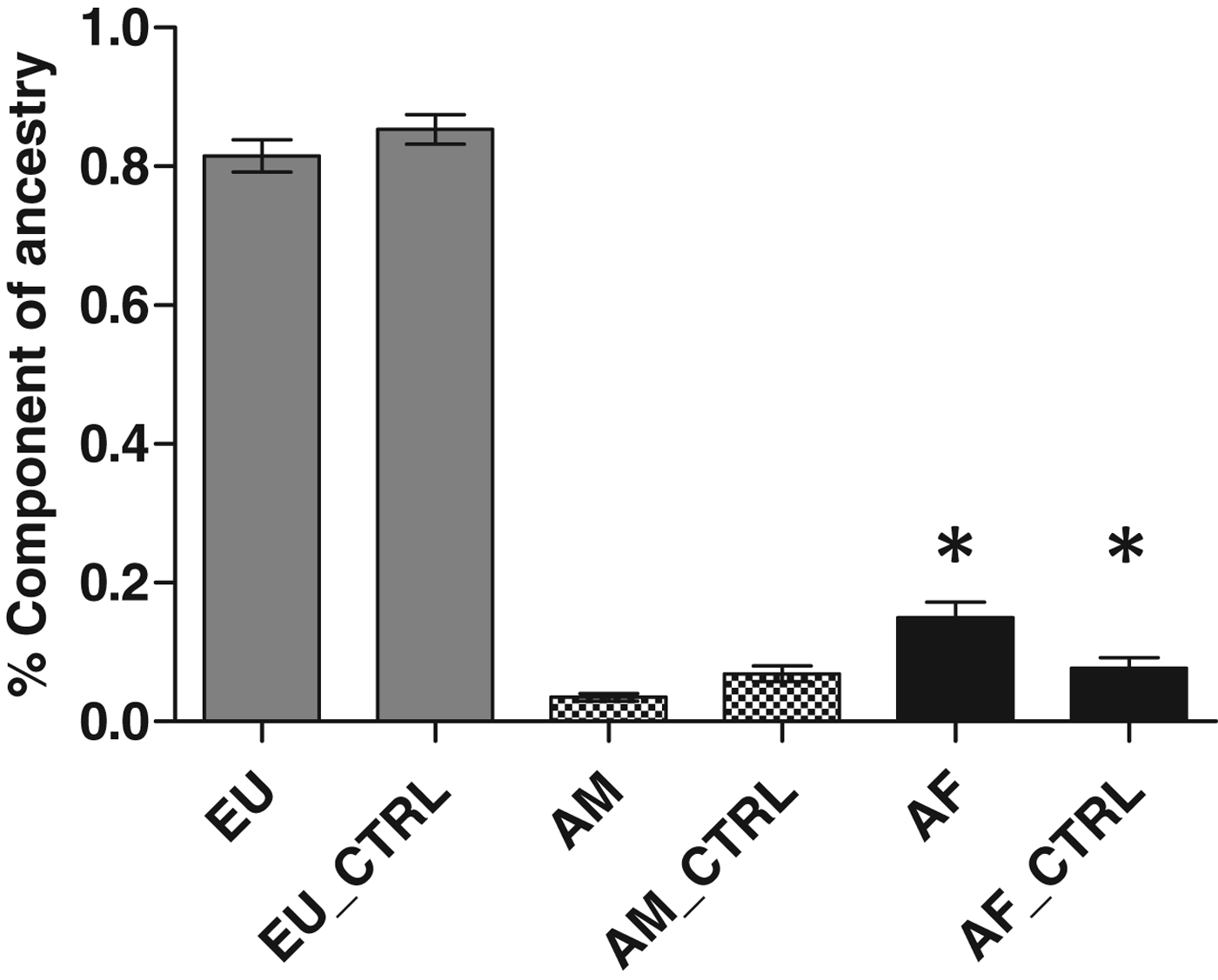
Fig. 1. Analysis of genomic ancestry of patients with lung cancer. EU, Europeans; AF, African; AM, Amerindians. Significant difference was found between Africans when compared with control group (P = 0·004).
4. Discussion
In the present study that focused on Brazilian lung cancer patients, the EGFR 746_750del, a mutation that is associated with a better response of NSCLC patients to TKIs, such as Gefitinib and Erlotinib (Han et al., Reference Han, Kim, Jeon, Hwang, Im, Lee, Kim, Kim, Heo, Kim, Chung and Bang2006; Riely et al., Reference Riely, Politi, Miller and Pao2006; Irmer et al., Reference Irmer, Funk and Blaukat2007) was detected somatically in only three of 88 tumour samples (3·4%). This is a significantly lower rate than the rates reported for ethnically diverse populations worldwide, with rates ranging from 15 to 40% of NSCLC analysed, primarily adenocarcinomas (Irmer et al., Reference Irmer, Funk and Blaukat2007; Matsuo et al., Reference Matsuo, Ito, Yatabe, Hiraki, Hirose, Wakai, Kosaka, Suzuki, Tajima and Mitsudomi2007; Leidner et al., Reference Leidner, Fu, Clifford, Hamdan, Jin, Eisenberg, Boggon, Skokan, Franklin, Cappuzzo, Hirsch, Varella-Garcia and Halmos2009; Tapia et al., Reference Tapia, Savic, Bihl, Rufle, Zlobec, Terracciano and Bubendorf2009; Vlastos et al., Reference Vlastos, Zinszner, Hussenet, du Manoir, Vordonis, Nikolakopoulou, Hardavella, Lacomme, Vignaud and Martinet2010). A previous study of Latin American patients from biopsies taken from Argentinean, Colombian, Peruvian and Mexican lung cancer patients (Arrieta et al., Reference Arrieta, Cardona, Bramuglia, Gallo, Campos-Parra, Serrano, Castro, Avilés, Amorin, Kirchuk, Cuello, Borbolla, Riemersma, Becerra and Rosell2011) reported the same deletion in 48·4% patients (185/382 cases) and this was in agreement with data from Asian (60%) and European populations (62·2%) (Rosell et al., Reference Rosell, Moran, Queralt, Porta, Cardenal, Camps, Majem, Lopez-Vivanco, Isla, Provencio, Insa, Massuti, Gonzalez-Larriba, Paz-Ares, Bover, Garcia-Campelo, Moreno, Catot, Rolfo, Reguart, Palmero, Sánchez, Bastus, Mayo, Bertran-Alamillo, Molina, Sanchez and Taron2009). A previous study also detected a low incidence of mutations in a Caucasian population (12%) and suggested that the specific lung cancer subtype analysed as well as the technique used could explain the wide range of somatic mutations reported (Wislez et al., Reference Wislez, Antoine, Baudrin, Poulot, Neuville, Pradere, Longchampt, Isaac-Sibille, Lebitasy and Cadranel2010). The L858R EGFR somatic mutation was found in only one patient and other mutations such as L861Q and G719S were not noted in the present study. These results are in line with data reported by Vlastos et al. (Reference Vlastos, Zinszner, Hussenet, du Manoir, Vordonis, Nikolakopoulou, Hardavella, Lacomme, Vignaud and Martinet2010) that analysed Caucasian population, but contrasts other studies reporting high rates of these EGFR mutations in NSCLC in patients of Asian (40%) and European (37·8%) descent (Han et al., Reference Han, Kim, Jeon, Hwang, Im, Lee, Kim, Kim, Heo, Kim, Chung and Bang2006; Murray et al., Reference Murray, Timotheadou, Linardou, Vrettou, Kostopoulos, Skrickova, Papakostantinou, Christodoulou, Pectasides, Samantas, Papakostas, Skarlos, Kosmidis and Fountzilas2006; Riely et al., Reference Riely, Politi, Miller and Pao2006; Irmer et al., Reference Irmer, Funk and Blaukat2007; Matsuo et al., Reference Matsuo, Ito, Yatabe, Hiraki, Hirose, Wakai, Kosaka, Suzuki, Tajima and Mitsudomi2007; Rosell et al., Reference Rosell, Moran, Queralt, Porta, Cardenal, Camps, Majem, Lopez-Vivanco, Isla, Provencio, Insa, Massuti, Gonzalez-Larriba, Paz-Ares, Bover, Garcia-Campelo, Moreno, Catot, Rolfo, Reguart, Palmero, Sánchez, Bastus, Mayo, Bertran-Alamillo, Molina, Sanchez and Taron2009). Specifically, the L858R mutation that has been reportedly detected in 12·5 to 45% of NSCLC-associated EGFR mutations (Pan et al., Reference Pan, Pao and Ladanyi2005; Tapia et al., Reference Tapia, Savic, Bihl, Rufle, Zlobec, Terracciano and Bubendorf2009) was found in only one case (1·1%) in the present study. This patient has features more common to NSCLC patients with the L585R mutation in exon 21, i.e. female, non-smoker with adenocarcinoma histology (Kondo et al., Reference Kondo, Yokoyama, Fukui, Yoshioka, Yokoi, Nagasaka, Imaizumi, Kume, Hasegawa, Shimokata and Sekido2005; Shigematsu et al., Reference Shigematsu, Lin, Takahashi, Nomura, Suzuki, Wistuba, Fong, Lee, Toyooka, Shimizu, Fujisawa, Feng, Roth, Herz, Minna and Gazdar2005; Riely et al., Reference Riely, Politi, Miller and Pao2006). One plausible explanation for the disparities between the rates of the EGFR mutations in the present study and other studies is the type of tumours and patients analysed. The cases analysed in the present study are mostly men, smokers, with a high African ancestry. Other studies showed that the prevalence of EGFR mutations among African Americans have yielded conflicting results (Cote et al., Reference Cote, Haddad and Edwards2011; Reinersman et al., Reference Reinersman, Johnson, Riely, Chitale, Nicastri, Soff, Schwartz, Sima, Ayalew, Lau, Zakowski, Rusch, Ladanyi and Kris2011). Indeed, in a previous study focusing on Brazilian NSCLC cases, Amoedo et al. (Reference Amoedo, Castelo-Branco, Paschoal, Pezzuto, Esperança, Rumjanek and Rumjanek2009) studied only exon 19 of EGFR and reported one 746_750del in 64 paraffin-embedded tissue samples analysed (∼1·5%). These data combined with the data presented herein support the notion of a low prevalence of somatic EGFR mutations in Brazilian NSCLC cases.
Somatic activating KRAS mutations in NSCLC can usually be detected in codon 12 (Gly12Asp), less frequently in codon 13 (Gly13Asp) and rarely at codon 61 (Gln61His) (Riely et al., Reference Riely, Marks and Pao2009). The frequency and type of KRAS activating mutations in NSCLC are in part determined by the specific tumour histology, patients’ ethnicity and smoking history (Riely et al., Reference Riely, Kris, Rosenbaum, Marks, Li, Chitale, Nafa, Riedel, Hsu, Pao, Miller and Ladanyi2008; Amoedo et al., Reference Amoedo, Castelo-Branco, Paschoal, Pezzuto, Esperança, Rumjanek and Rumjanek2009); these mutations are more commonly encountered in Caucasians, smokers with adenocarcinoma histology (Suda et al., Reference Suda, Tomizawa and Mitsudomi2010). In the present study, the mutation in codon 12 was noted in male patients diagnosed with adenocarcinoma. Unlike the reported mutational spectrum in the KRAS gene in other world populations (Dearden et al., Reference Dearden, Stevens, Wu and Blowers2013), the sequence alteration described in codon 61 was detected in two female smokers, diagnosed with SCC. Although environmental factors could be involved, the aetiology of this alteration remains to be established.
The rate of somatic KRAS mutations in the present study was low (7·9%), similar to the low rates (11/173; 6·3%) reported by Lee et al. (Reference Lee, Kim, Jin, Yoo, Park, Choi, Jeon, Cho, Lee, Cha, Park, Kim, Jung and Park2010) who analysed 173 Korean cases. These rates are well below the rates reported for Japanese (12·8%) (Lee et al., Reference Lee, Kim, Jin, Yoo, Park, Choi, Jeon, Cho, Lee, Cha, Park, Kim, Jung and Park2010), Latin American (16·6%) (Arrieta et al., Reference Arrieta, Cardona, Bramuglia, Gallo, Campos-Parra, Serrano, Castro, Avilés, Amorin, Kirchuk, Cuello, Borbolla, Riemersma, Becerra and Rosell2011) and Caucasian populations (41·9%) (Borràs et al., Reference Borràs, Jurado, Hernan, Gamundi, Dias, Martí, Mañé, Arcusa, Agúndez, Blanca and Carballo2011). These apparent differences in somatic mutation rates in the KRAS gene may also be attributed to different ethnic background of the studied populations and may indicate that the malignant transformation process in Brazilian NSCLC cases may have different pathways than those in some other, ethnically diverse, populations.
Although several studies have shown the relationship of BRAF and PTEN with the tumourigenesis of lung cancer, several studies confirm the low frequency of mutations of these genes in NSCLC (Forgacs et al., Reference Forgacs, Biesterveld, Sekido, Fong, Muneer, Wistuba, Milchgrub, Brezinschek, Virmani, Gazdar and Minna1998; Yokomizo et al., Reference Yokomizo, Tindall, Drabkin, Gemmill, Franklin, Yang, Sugio, Smith and Liu1998; Sasaki et al., Reference Sasaki, Kawano, Endo, Suzuki, Haneda, Yukiue, Kobayashi, Yano and Fujii2006; Pratilas et al., Reference Pratilas, Hanrahan, Halilovic, Persaud, Soh, Chitale, Shigematsu, Yamamoto, Sawai, Janakiraman, Taylor, Pao, Toyooka, Ladanyi, Gazdar, Rosen and Solit2008; Jin et al., Reference Jin, Kim, Jeon, Choi, Kim, Lee, Cha, Yoon, Kim, Jung and Park2010; Marchetti et al., Reference Marchetti, Felicioni, Malatesta, Grazia Sciarrotta, Guetti, Chella, Viola, Pullara, Mucilli and Buttitta2011). In the Brazilian population analysed here, we have not found any mutations in BRAF and EGFR genes thus confirming the rarity of this alteration in NSCLC. To our knowledge, our work represents the first data of a PTEN and BRAF V600E mutation search in Brazilian patients diagnosed with NSCLC.
Approximately 80% of adenocarcinomas express TTF-1, and this marker has been used to differentiate the histological subtypes of NSCLC (adenocarcinoma and SCC) (Maeshima et al., Reference Maeshima, Omatsu, Tsuta, Asamura and Matsuno2008). Our results showed that 40·7% of SCCs analysed were TTF-1 (+) demonstrating that a subset of SCCs should be considered NSCLC-favour adenocarcinoma (Travis et al., Reference Travis, Brambilla and Riely2013). According to Ordóñez (Reference Ordóñez2012), the introduction of target therapies can result in dramatically different outcomes based on histological subtype. Thus, the use of markers such as TTF-1 can help to discriminate between adenocarcinoma and SCC subtypes, what is indispensable to define a personalized treatment.
The Brazilian population has a major Caucasian contribution (Pena et al., Reference Pena, Bastos-Rodrigues, Pimenta and Bydlowski2009) but our study found a greater African component in patients with NSCLC than in the control group. African-American patients with NSCLC are significantly less likely to harbour activating mutations in EGFR genes and their signalling pathway when compared with Caucasians (Riely et al., Reference Riely, Politi, Miller and Pao2006). This notion is further supported by Reinersman et al. (Reference Reinersman, Johnson, Riely, Chitale, Nicastri, Soff, Schwartz, Sima, Ayalew, Lau, Zakowski, Rusch, Ladanyi and Kris2011), who reported that African Americans are less likely to have KRAS mutations in NSCLC when compared with Caucasians, a finding corroborated in our study.
As demonstrated, our data show a low frequency of mutations in EGFR, KRAS, BRAF and PTEN. This may be related to the high African component found in our patients, which has been discussed in previous studies (Maxwell et al., Reference Maxwell, Risinger, Hayes, Alvarez, Dodge, Barrett and Berchuck2000; Kumar et al., Reference Kumar, Brim, Giardiello, Smoot, Nouraie, Lee and Ashktorab2009; Leidner et al., Reference Leidner, Fu, Clifford, Hamdan, Jin, Eisenberg, Boggon, Skokan, Franklin, Cappuzzo, Hirsch, Varella-Garcia and Halmos2009; Pena et al., Reference Pena, Bastos-Rodrigues, Pimenta and Bydlowski2009; Reinersman et al., Reference Reinersman, Johnson, Riely, Chitale, Nicastri, Soff, Schwartz, Sima, Ayalew, Lau, Zakowski, Rusch, Ladanyi and Kris2011) but had not yet been analysed in the Brazilian population. Thus, our data add to a growing body of evidence that highlight the genetic heterogeneity of the EGFR pathway in NSCLC among different populations, which highlights the need to incorporate these differences in designing clinical therapies and agents for the inhibition of this pathway.
The limitations of the present study should be pointed out: this is a relatively small study from a single medical centre that serves a population that might not reflect the ethnic diversity of the entire Brazilian population. In addition, it must be said that patients are referred to treatment when surgical procedures are no longer acceptable and further investigations inappropriate. Hence it is important to stress that these results should be expanded and validated in larger studies focusing on Brazilian patients.
In conclusion, Brazilian lung cancer patients display a low frequency of somatic mutations in EGFR, KRAS, BRAF and PTEN. This may be related to the high African ancestry component found in our patients, but an expansion and validation of these preliminary data is needed.
Acknowledgements
We are grateful to all patients.
Financial Support
This work was partially supported by FAPEMIG; CNPq and INCT em Medicina Molecular; Brasil.
Statement of Interest
None.






