1. Introduction
Heterosis, or hybrid vigour, has been widely used in genetic crop improvement, and the genetic basis of heterosis has also been investigated. Dominance (Davenport, Reference Davenport1908) and over-dominance (Shull, Reference Shull1908) are two hypotheses that were proposed a century ago to explain the genetic basis of heterosis. Recent advances in genome research involving a number of molecular-marker techniques and the availability of high-density molecular linkage maps, together with developments in analytical methods (Zeng, Reference Zeng1994), facilitated the analysis of the genetic basis of quantitative traits. Many quantitative trait locus (QTL) mapping studies have provided insight into the genetic basis of heterosis (Xiao et al., Reference Xiao, Li, Yuan and Tanksley1995; Li et al., Reference Li, Luo, Mei, Shu, Tablen, Zhong, Ying, Stansel, Khush and Paterson2001, Reference Li, Lu, Chen, Mu, Hu and Li2008; Hua et al., Reference Hua, Xing, Wu, Xu, Sun, Yu and Zhang2003; Melchinger et al., Reference Melchinger, Utz and Schön2008; Luo et al., Reference Luo, Fu, Zhang, Wu, Tian, Liu, Zhu, Yang and Sun2009a), but reached different conclusions. One known problem in establishing the genetic basis of heterosis is the use of whole-genome segregating populations, where interactions often mask the effects of individual loci (Semel et al., Reference Semel, Nissenbaum, Menda, Zinder, Krieger, Issman, Pleban, Lippman, Gur and Zamir2006).
Introgression lines (ILs) are the results of using marker-assisted selection (MAS) to introgress small chromosomal segments from the donor into the recurrent parent by consecutive backcrossing and selfing (Eshed & Zamir, Reference Eshed and Zamir1994). Any phenotypic difference between such an IL and its recurrent parent should be due to QTLs located on the introgressed segments from the donor. Consequently, ILs are a more precise estimate of the genetic effects of introgression under a relatively uniform and elite lineage background (Tanksley & Nelson, Reference Tanksley and Nelson1996). They are, therefore, well suited for use in the genetic analysis of heterosis.
Recently, we reported QTL analysis of panicle-related traits and identified heterotic loci (HLs) associated with six yield-related traits in a set of 265 ILs (Luo et al., Reference Luo, Tian, Fu, Yang and Sun2009b). The lines were generated from a cross between Guichao 2, a high-yield commercial Indica cultivar (Oryza sativa L.), as the recurrent parent, and a common wild rice accession collected from Dongxiang County, Jiangxi Province, China, as the donor parent. In our previous study, the mid-parental heterosis (H MP) values were calculated as H MP=F1–(IL+Guichao 2)/2. The H MP values were used to identify loci affecting heterosis in the six yield-related traits. An HL is determined when a locus demonstrates a significant difference between the heterozygote and the mean of the two corresponding homozygotes; that is, the HL is a QTL for heterosis (Luo et al., Reference Luo, Wu, Tian, Za, Dong, Fu, Wang, Yang and Sun2011).
In this study, based on the set of 265 ILs and 265 testcross F1s (derived from the ILs and the recurrent parent, Guichao 2), QTLs and HLs associated with yield-related traits in testcross F1s were analysed. The genetic effects and main features of the HLs were discussed.
2. Materials and methods
(i) Experimental population and field trials
The IL population comprised 265 lines carrying variant introgressed segments of Dongxiang common wild rice (Oryza rufipogon) collected from Dongxiang county, Jiangxi Province, in the background of an Indica (O. sativa L. ssp. indica) cultivar, Guichao 2. The F1 population comprised 265 testcrosses derived from crosses between the 265 ILs and the recurrent parent Guichao 2. These 265 lines represented 81·5% of the O. rufipogon genome. The detailed characteristics of the ILs were presented in Luo et al. (Reference Luo, Tian, Fu, Yang and Sun2009b, Reference Luo, Wu, Tian, Za, Dong, Fu, Wang, Yang and Sun2011). The F1 testcross individuals and the corresponding parental ILs were evaluated in the summer of 2004 at the Experiment Station of the China Agricultural University (ESCAU), Beijing (39°N, 116°E).
Field trials of the 265 ILs, the 265 testcross F1s and the recurrent parent Guichao 2 were conducted at Beijing–ESCAU in summer 2004. The detailed field trials were presented in Luo et al. (Reference Luo, Wu, Tian, Za, Dong, Fu, Wang, Yang and Sun2011) . Ten plants from each line were harvested at maturity in both the IL and F1 populations, and the following traits were scored: the number of spikelets per panicle (SP), the number of filled grains per panicle (GP), per cent seed set (SSP), 1000-grain weight (GW), the number of panicles per plant (PP) and grain yield per plant (YP).
(ii) Data analysis
The simple sequence repeat (SSR) markers analysed in this study were taken from previous publications (McCouch et al., Reference McCouch, Teytelman, Xu, Lobos, Clare, Walton, Fu, Maghirang, Li, Xing, Zhang, Kono, Yano, Fjellstrom, DeClerck, Schneider, Cartinhour, Ware and Stein2002). The detailed characteristics of the SSR markers in the ILs were presented in Luo et al. (Reference Luo, Tian, Fu, Yang and Sun2009b) . A total of 160 polymorphic SSR markers were used to genotype the 265 ILs and the recurrent parent Guichao 2, following Tian et al. (Reference Tian, Li, Fu, Zhu, Fu, Wang and Sun2006) . The F1 testcross genotypes were deduced based on the genotype of their corresponding parental ILs. The direct trait measurement values from the six yield-related traits obtained from the ILs were used to identify the associated QTLs. The testcross F1 trait measurements were used to identify loci affecting testcross F1 performance.
Based on IL structure, QTLs can be mapped on introgressed chromosome segments. One representative marker for each specific introgressed segment was defined as a QTL (Luo et al., Reference Luo, Wu, Tian, Za, Dong, Fu, Wang, Yang and Sun2011). The association between the phenotype and 160 SSR marker data was investigated by single-point analysis using the software package Map Manager QTXb17 (Manly et al., Reference Manly, Cudmore and Meer2001). The statistical a priori threshold for main effect loci was P<0·01 (the probability that loci had no effect on the trait).
The genetic effects in the testcross F1s were defined as follows: d=H MP=[F1–(IL+Guichao 2)/2] (Luo et al., Reference Luo, Wu, Tian, Za, Dong, Fu, Wang, Yang and Sun2011); the trait mean values in the testcross F1s were ![]() , where a is the additive effects from the performance values of testcross F1; IL is the mean value for the same measured trait in the corresponding IL parent, and the homozygous IL genotype value was 2a; here Guichao 2 genotype value was assumed as zero for a simple case; subsequently HL effects were inferred by comparisons between the genetic effects on F1 performance and H MP. HLs with the ratio between dominant and additive effects d/a⩽1 were considered complete or partial dominant loci, and expected to generate an estimate of F1 performance (a+d) equal to or higher than twice the H MP (d). HLs with d/a>1, that is, 2d (2×H MP)>a+d (F1), or only detectable for H MP, were determined as over-dominant loci (Melchinger et al., Reference Melchinger, Utz and Schön1998).
, where a is the additive effects from the performance values of testcross F1; IL is the mean value for the same measured trait in the corresponding IL parent, and the homozygous IL genotype value was 2a; here Guichao 2 genotype value was assumed as zero for a simple case; subsequently HL effects were inferred by comparisons between the genetic effects on F1 performance and H MP. HLs with the ratio between dominant and additive effects d/a⩽1 were considered complete or partial dominant loci, and expected to generate an estimate of F1 performance (a+d) equal to or higher than twice the H MP (d). HLs with d/a>1, that is, 2d (2×H MP)>a+d (F1), or only detectable for H MP, were determined as over-dominant loci (Melchinger et al., Reference Melchinger, Utz and Schön1998).
3. Results
(i) The relationships among the mean trait values of ILs, HMP, and F1 performance
The correlation coefficients between testcross F1 mean values, H MP values and parental IL mean values for yield-related traits are shown in Table 1. In general, a positive but lower correlation between IL trait values and the F1s was observed; the average R2 (determination coefficients) was 0·235, which suggested that additive gene action made a small contribution to F1 performance. A general negative correlation trend was evident between IL and H MP trait values, clearly suggesting that additive and dominant gene action operated independently in the testcross population. In the 265 testcross F1s, a highly positive correlation between F1 performance and H MP values was observed for all traits, with an average R2 =0·580 (range of 0·428 for GW to 0·775 for YP), indicating that H MP largely influenced F1 performance.
Table 1. Phenotypic correlation (R) and determination coefficients (R2) for six yield-related traits between IL and testcross F1 performance values and H MP values
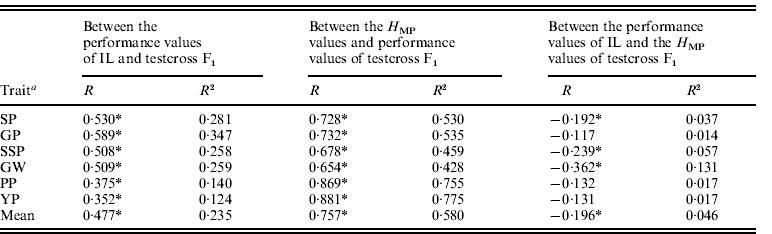
a Trait abbreviations: the number of spikelets per panicle (SP), the number of filled grains per panicle (GP), per cent seed set (SSP), 1000-grain weight (GW), the number of panicles per plant (PP) and grain yield per plant (YP).
* Significance levels P<0·01.
(ii) Genetic effects of QTLs in F1 testcross population
IL phenotypic data from six yield-related traits were used to identify the associated QTLs. Fifty-four QTLs were detected (partial QTLs detected in ILs are indicated in underlined text in Table 2). Trait phenotypic values from F1 testcrosses were used to infer the QTLs contributing to F1 testcross performance. Fifty-one QTLs influencing F1 testcross performance were detected for the six yield-related traits (partial QTLs detected in the F1 testcrosses are shown in Table 2). In our previous study, the H MP values in F1 testcross were used to infer which QTLs contributed to heterosis. A total of 42 H MP QTLs (or HLs) associated with H MP values were detected for the six yield-related traits (Luo et al., Reference Luo, Wu, Tian, Za, Dong, Fu, Wang, Yang and Sun2011, all H MP QTLs are shown in Table 2).
Table 2. The genetic effects of HLs on six yield-related traits in the testcross F1s (the HLs were mapped by Luo et al., Reference Luo, Wu, Tian, Za, Dong, Fu, Wang, Yang and Sun2011)
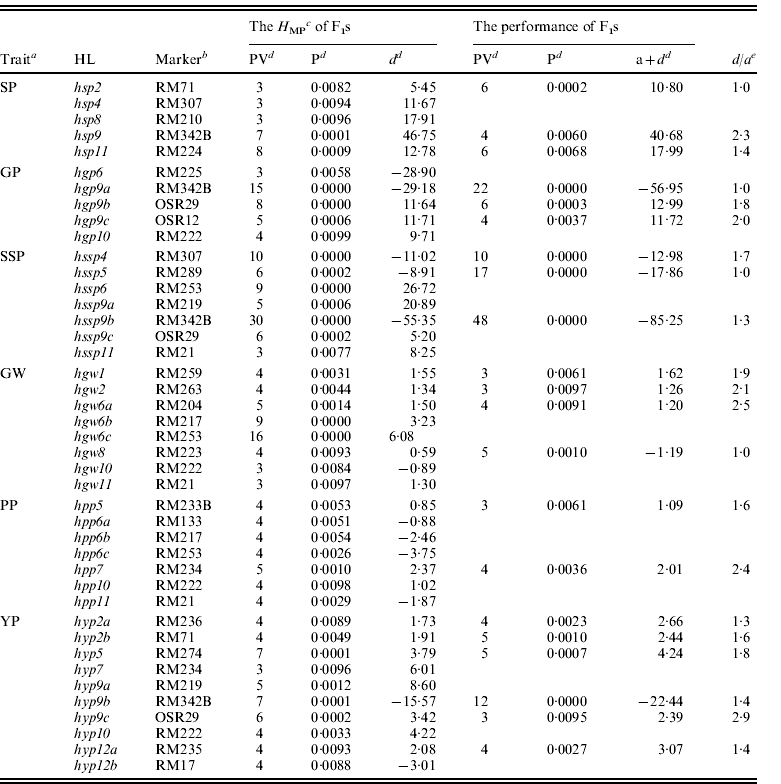
a See Table 1 for abbreviations.
b Markers indicated in underlined text are QTLs identified in ILs.
c H MP is the mid-parental heterosis of testcross F1.
d PV, the phenotypic variance explained by the locus; P, the probability that the marker genotype had no effect on the trait; a+d, the additive and dominance effects from the performance values of testcross F1; d, the dominance effect from the H MP values.
e d/a, the ratio between dominant and additive effects.
Table 2 indicates the genetic overlap of H MP and QTLs detected in the F1 testcrosses. Of 42 H MP QTLs, 21 loci were only associated with H MP, and showed over-dominant expression. The other 21 loci simultaneously influenced H MP and F1 performance. A comparison of the genetic effects of loci detected in both H MP and F1 testcross performance indicated a d/a⩽1 in hsp2, hgp9a, hssp5 and hgw8, suggesting dominant loci, and over-dominant effects in the remaining 17 loci (d/a>1). In 42 of the HLs, 38 (90·5%) were over-dominant and four appeared dominant. These results indicated that at the single locus level, HLs were predominantly over-dominant. In IL QTL analysis, nine (21·4%) of the above 42 HLs were resolved at the same statistical threshold, and showed less genetic overlap with the six yield-related trait QTLs. These results are consistent with a lower correlation between IL trait values and the corresponding F1s.
4. Discussion
The complex nature of heterosis makes it difficult to partition it into individual components, particularly in F2, backcrossed, and recombinant inbred populations; the epistatic interactions among the many segregating loci throughout the genome makes it difficult to define specific heterotic phenotypes and the individual genomic loci that control them (Li et al., Reference Li, Luo, Mei, Shu, Tablen, Zhong, Ying, Stansel, Khush and Paterson2001; Semel et al., Reference Semel, Nissenbaum, Menda, Zinder, Krieger, Issman, Pleban, Lippman, Gur and Zamir2006; Xin et al., Reference Xin, Wang, Xin, Wang, Yang and Luo2011). To overcome these limitations, we developed a set of ILs carrying a few chromosome segments from the wild rice species O. rufipogon (Luo et al., Reference Luo, Tian, Fu, Yang and Sun2009b). This IL population allowed us to partition heterosis into defined genomic regions, eliminating a major part of the genome-wide epistasis. The heterotic effects were determined as the combined effects of both additive and dominant gene actions, estimated from the performance values of testcross F1s and the dominance effects estimated from the H MP values of testcross F1s. Based on this strategy, we characterized the gene action type at each HL. Forty-two HLs for six yield-related traits revealed two different genetic effects: dominance or over-dominance. These HL data indicated that over-dominance was the major underlying factor of heterosis. Thirty-eight (90·5%) HLs exhibited over-dominant effects and only four HLs showed dominant effects. Notably, Semel et al. (Reference Semel, Nissenbaum, Menda, Zinder, Krieger, Issman, Pleban, Lippman, Gur and Zamir2006) carried out quantitative genetic and phenotypic analyses on an IL population of tomato (Solanum lycopersicum) carrying a single chromosomal segment from the distantly related wild species Solanum pennellii. That study generated results congruent with the present study; in the absence of epistasis, at a single locus level, over-dominant loci had greater effects on tomato yield and fitness.
The exploitation of favourable genes from wild rice might further improve tolerance to biotic and abiotic stress, yield and other important agronomic traits for rice variety. Luo et al. (Reference Luo, Wu, Tian, Za, Dong, Fu, Wang, Yang and Sun2011) investigated the HLs derived from wild rice, thought that favourable HLs capable of improving agronomic traits are available, and indicated that the identification of HLs between wild rice and cultivated rice could lead to a new strategy for the application of heterosis in rice breeding. It is generally known that the over-dominant HLs is more advantageous than dominant HLs in heterosis utilization. In this study, 28 (66·7%) of 42 HLs showed significantly positive over-dominant effects (P<0·01) on yield-related traits, suggesting that these markers are viable candidates for marker-aided improvement of rice yield potential.
Previous study (Li et al., Reference Li, Lu, Chen, Mu, Hu and Li2001) revealed that backcross F1 performance was largely determined by dominant gene action. Mei et al. (Reference Mei, Li, Shu, Guo, Wang, Yu, Ying and Luo2005) analysed the correlation between RILs and backcrossed populations for agricultural traits, and considered backcross F1 performance was mainly determined by non-additive gene action. Our study employed a similar experimental design and found a highly positive correlation between testcross F1 and H MP F1 testcross performance values, and a lower positive correlation between IL performance values and F1 testcross performance values (Table 1). These results indicated that dominance gene action rather than additive gene action was a substantial contributor to F1 testcross performance. Furthermore, the testcross described in this study corresponds to previous backcross studies (Li et al., Reference Li, Luo, Mei, Shu, Tablen, Zhong, Ying, Stansel, Khush and Paterson2001; Mei et al., Reference Mei, Li, Shu, Guo, Wang, Yu, Ying and Luo2005). The negative correlation between IL performance values and H MP values of the F1 testcross population clearly indicated that additive and dominant gene action acted independently in the testcross population. QTL and HL analyses demonstrated that nine of the 42 HLs were also detected in the QTL analysis (Table 2), and exhibited less genetic overlap with QTLs, consistent with results reported by Hua et al. (Reference Hua, Xing, Wu, Xu, Sun, Yu and Zhang2003) . Therefore, heterosis and trait performance may be conditioned by different sets of loci.
We thank the anonymous referees for their critical comments on this manuscript. This research was supported by a grant (no. 2007CB109002) from the National Basic Research Program of China (973 program).




