1. Introduction
Many plants have genetically controlled mechanisms to promote outcrossing (self-incompatibility or SI systems), which are thought to evolve under conditions of high inbreeding depression and to result in elevated genetic load through increased heterozygosity (Charlesworth & Charlesworth, Reference Charlesworth and Charlesworth1979a; Charlesworth et al., Reference Charlesworth, Morgan and Charlesworth1990; Byers & Meagher, Reference Byers and Meagher1992; Charlesworth, Reference Charlesworth2006b). There has thus been an expectation that loss of SI should have detrimental fitness consequences, due to the unmasking of deleterious mutations resulting from increased homozygosity and loss of genetic diversity. The role of inbreeding depression in maintaining outcrossing has received thorough theoretical evaluation (Charlesworth & Charlesworth, Reference Charlesworth and Charlesworth1979b; Lande & Schemske, Reference Lande and Schemske1985; Charlesworth & Charlesworth, Reference Charlesworth and Charlesworth1987; Barrett, Reference Barrett1988; Charlesworth & Charlesworth, Reference Charlesworth and Charlesworth1990; Barrett & Charlesworth, Reference Barrett and Charlesworth1991; Uyenoyama & Waller, Reference Uyenoyama and Waller1991; Charlesworth et al., Reference Charlesworth, Morgan and Charlesworth1992a; Husband & Schemske, Reference Husband and Schemske1996; Charlesworth & Charlesworth, Reference Charlesworth and Charlesworth1999; Wang et al., Reference Wang, Hill, Charlesworth and Charlesworth1999; Carr & Dudash, Reference Carr and Dudash2003) and has been evaluated empirically since the time of Darwin (Darwin, Reference Darwin1876).
Strongly SI species should benefit from sheltering of the genetic load but can suffer from the cost of a reduced number of compatible mating partners compared with selfing individuals (i.e. reproductive assurance), particular in small populations where the number of different mating types might be low. SI is a frequent evolutionary transition at the level of species and can also occur within species (Weller & Sakai, Reference Weller and Sakai1999), suggesting costs to SI or advantages of selfing under certain conditions. Shifts to self-compatibility are often considered to be unidirectional due to the difficulty of re-establishing a functional SI system (Igic et al., Reference Igic, Bohs and Kohn2006). However, intermediate levels of self-fertilization can be maintained within species that do not have functional SI systems (Lloyd, Reference Lloyd1979; reviewed in Goodwillie et al., Reference Goodwillie, Kalisz and Eckert2005) and reversion to outcrossing theoretically could be achieved by other mechanisms. Although theory predicts that intermediate rates of outcrossing should be unstable (Lande & Schemske, Reference Lande and Schemske1985), observed patterns suggest that this might not always be true (reviewed in Goodwillie et al., Reference Goodwillie, Kalisz and Eckert2005). A balance somewhere between complete outcrossing and complete inbreeding could be driven by a dynamic trade-off between inbreeding for reproductive assurance or colonization ability and outcrossing for offspring quality (Bateman, Reference Bateman1955; Jarne & Charlesworth, Reference Jarne and Charlesworth1993; Charlesworth, Reference Charlesworth2006b).
The realized consequences of inbreeding also depend on the particular history of outcrossing in a species, population or family. Theoretical models predict that inbreeding depression should evolve with selfing due to exposure of deleterious recessive mutations (Schemske & Lande, Reference Schemske and Lande1985), but should eventually end up being lower in selfing than in outcrossing populations due to purging of deleterious recessive mutations. However, the strength of this effect depends on factors such as the magnitude and duration of inbreeding, the genetic basis of inbreeding depression (i.e. partial dominance vs overdominance), the number of loci that contribute, the magnitude of effects of alleles at these loci, linkage to genes under viability selection, population size and the developmental stage at which inbreeding depression acts (Charlesworth & Charlesworth, Reference Charlesworth and Charlesworth1987; Barrett & Charlesworth, Reference Barrett and Charlesworth1991; Charlesworth et al., Reference Charlesworth, Morgan and Charlesworth1992; Husband & Schemske, Reference Husband and Schemske1996). For example, if recessive mutations have only a mildly deleterious effect or a low dominance coefficient, effective purging will not occur and substantial inbreeding depression could be maintained even in highly inbred populations (Charlesworth et al., Reference Charlesworth, Morgan and Charlesworth1990, Reference Charlesworth, Morgan and Charlesworth1991).
The expected impact of inbreeding is also complicated by such factors as population subdivision and biogeographical history (Vekemans et al., Reference Vekemans, Schierup and Christianssen1998; Schierup et al., Reference Schierup, Vekemans and Charlesworth2000; Charlesworth, Reference Charlesworth2003), as well as life history (Morgan, Reference Morgan2001) and local environmental effects (Hayes et al., Reference Hayes, Winsor and Stephenson2005). For example, gene flow between populations with different mating systems could affect the rate at which purging might occur. Particularly for conservation purposes, there has been much discussion of whether purging of genetic load can mitigate the detrimental effects of reduced heterozygosity caused by population fragmentation and over what time scales this might occur (Hedrick & Kalinowski, Reference Hedrick and Kalinowski2000; Glémin et al., Reference Glémin, Bataillon, Ronfort, Mignot and Olivieri2001; Keller & Waller, Reference Keller and Waller2002). Although empirical evidence remains equivocal, it has been suggested that plant populations with slight inbreeding and strong population structure should be the most likely to show purging (reviewed in Keller & Waller, Reference Keller and Waller2002). Local adaptation of inbred populations could also result in outcrossing depression (Bailey & McCauley, Reference Bailey and McCauley2006), which could seriously affect the balance between selfing and outcrossing, even in the face of some inbreeding depression. Probably due to the difficulty of forcing selfing in strongly SI species, most data on purging come from studies on populations or species that do not have genetically controlled mechanisms to ensure outcrossing, where the fitness of selfed and outcrossed progeny can be directly compared. Since SI species normally will have experienced a long period of outcrossing, the magnitude of inbreeding depression is expected to be high and at least some degree of purging is probably necessary in order for effective shifts to inbreeding to occur.
In order to understand the selective forces at work in mating system regulation, one approach is to compare the genetic consequences of inbreeding in closely related species that are predicted to have shared the same self-incompatibility system in the past. Selfing is expected to reduce both effective population size and effective recombination rates (N e), which result in reduced polymorphism, increased linkage disequilibrium and hitch-hiking between linked genes (Charlesworth & Wright, Reference Charlesworth and Wright2001; Wright et al., Reference Wright, Lauga and Charlesworth2002; Charlesworth, Reference Charlesworth2003; Glémin et al., Reference Glémin, Bazin and Charlesworth2006). Increased isolation between selfing populations relative to outcrossers can result directly from lack of outcrossing or indirectly from accompanying changes, such as small flowers and low pollen output (Glémin et al., Reference Glémin, Bazin and Charlesworth2006). The effects of population structure resulting from this isolation mean that it is not possible to make general predictions about the relative species-wide diversity in inbreeders and outcrossers, despite the reduced N e caused by homozygosity, because high levels of diversity can be maintained between selfing populations (Charlesworth, Reference Charlesworth2003). This means that selfing species could actually show higher levels of diversity than their outcrossing relatives.
Although the factors that result in loss of SI could be different from those that favour its evolution (Uyenoyama, Reference Uyenoyama1991), uncovering the mechanisms and fitness consequences of loss of SI could shed light on how selection is involved in the evolution of genetically controlled outcrossing mechanisms. Recent technological advances mean that it is now possible to consider such consequences at the whole-genome level and to test models of changes in rates of recombination and linkage disequilibrium in relation to mating systems (Charlesworth & Wright, Reference Charlesworth and Wright2001). Up to now, the self-compatible model plant Arabidopsis thaliana has been the main source of information for plant genomics. It is also in the same family as cultivated plants in the genus Brassica, where the mechanistic control of sporophytic SI has been extensively studied. It is thus understandable that recent studies have focused on SI in outcrossing relatives of A. thaliana, and the Brassicaceae make a logical system to focus on for understanding the genetic consequences of loss of SI. The pending complete genome sequences of Arabidopsis lyrata and Capsella rubella will greatly add to this potential.
The purpose of this paper is not to provide a comprehensive review but to highlight recent research in the Brassicaceae that demonstrates approaches to understanding the consequences and causes of loss of self-incompatibility. An overview of self-incompatibility in the Brassicaceae will first be presented, followed by a discussion of the genetic consequences of shifts to inbreeding in terms of mechanisms for loss of SI, changes in genetic diversity following loss of SI, and inbreeding depression in relation to outcrossing history.
2. SI in Brassicaceae
SI systems have long been of interest to both population geneticists interested in the spectacular levels of polymorphism resulting from the long times to coalescence of alleles involved in cell–cell recognition processes (thought to be maintained through the forces of balancing selection: Charlesworth, Reference Charlesworth1988; Charlesworth & Awadalla, Reference Charlesworth and Awadalla1998; Charlesworth et al., Reference Charlesworth, Awadalla, Mable and Schierup2000; Kamau & Charlesworth, Reference Kamau and Charlesworth2005), and by biochemists interested in the complex signalling pathways involved in mediating this type of response system (Nasrallah & Wallace, Reference Nasrallah and Wallace1967; Goring & Rothstein, Reference Goring and Rothstein1992; Stone et al., Reference Stone, Arnoldo and Goring1999; Nasrallah, Reference Nasrallah2000; Goring & Walker, Reference Goring and Walker2004). The principle for most types of SI reactions is that, if proteins on the surface of the pollen are recognized as self by proteins in the female receiving tissues (whether this occurs on the surface of the stigma, further down in the style or in the ovaries varies between types of SI), a signal is sent to block pollen tube growth. Since the determination of self involves independent genes in the male and female components, recognition specificity must be maintained between them. If this lock-and-key mechanism is disrupted, SI is expected to break down.
In sporophytic SI systems (SSI) characteristic of the Brassicaceae, genes coding for SI specificity in pollen (SCR for S-locus Cysteine Rich) and pistils (SRK for S-Related Kinase) are organized into self-recognition haplotypes (i.e. male and female genes are found in the same S-gene region) that can span over 100 kb (Suzuki et al., Reference Suzuki, Kai, Hirose, Fukui, Nishio and Takayama1999; Kusaba et al., Reference Kusaba, Dwyer, Hendershot, Vrebalov, Nasrallah and Nasrallah2001; Shiba et al., Reference Shiba, Kenmochi, Sugihara, Iwano, Kawasaki and Suzuki2003). It is thought that low recombination in these regions is essential to maintain the same specificity in male and female components (Awadalla & Charlesworth, Reference Awadalla and Charlesworth1999) but it is not known whether this would be maintained (or for how long) if SI functioning were lost.
In strongly SI individuals, lack of recombination in the S-gene region can lead to balancing selection extending to genes in the vicinity of those actually under selection (Charlesworth, Reference Charlesworth2006a). This is reflected in trans-specific polymorphisms in genes near the S-locus (Charlesworth et al., Reference Charlesworth, Kamau, Hagenblad and Tang2006) and high levels of linkage disequilibrium with genes flanking S-genes (Hagenblad et al., Reference Hagenblad, Bechsgaard and Charlesworth2006). Inbreeding populations are expected to experience decreased rates of recombination compared with their outcrossing relatives (reviewed in Charlesworth et al., Reference Charlesworth, Vekemans, Castric and Glemin2005) but if low rates of recombination are maintained by selection in functional SI haplotypes, the constraint to maintain coordination between male and female components might be relaxed in non-functional haplotypes (or in selfing populations). However, this assumes that S-genes would evolve neutrally following loss of SI because their only function is in mate recognition. Intriguingly, in a survey of the S-locus region in worldwide accessions of A. thaliana, although evidence of recombination at the S-locus was found, no evidence was found for recombination between the pseudogene orthologues of SRK and SCR (Sherman-Broyles et al., Reference Sherman-Broyles, Boggs, Farkas, Liu, Vrebalov and Nasrallah2007). Whether this is because loss of SI is too recent for a change to be observed, or whether some other selective force maintains the pairing, remains to be determined.
The name sporophytic SI comes from the finding that haploid pollen grains can carry a diploid complement of SCR proteins on their surface because they are deposited by diploid cells in the tapetum. Dominance interactions determine whether one or both SCR alleles from the sporophytic tissues are expressed on the surface of the pollen and which SRK proteins are presented to the pollen grain at the surface of the stigma (reviewed in Charlesworth et al., Reference Charlesworth, Awadalla, Mable and Schierup2000; Hatakeyama et al., Reference Hatakeyama, Takasaki, Suzuki, Nishio, Watanabe and Isogai2001). In addition, pollen and stigma can show different dominance for some haplotypes: for example, two haplotypes can be co-dominant in the stigma so that both are expressed, whereas one might be dominant over the other in pollen so that only one is expressed (Bateman, Reference Bateman1955; Thompson & Taylor, Reference Thompson and Taylor1966). This means that individuals sharing some S-haplotypes can produce viable offspring, which can effectively increase inbreeding levels even with a strong SI system. Dominance is thought to be under the control of SCR rather than SRK expression, as all SRK alleles appear to be expressed, regardless of dominance status (Hatakeyama et al., Reference Hatakeyama, Takasaki, Suzuki, Nishio, Watanabe and Isogai2001; Kusaba et al., Reference Kusaba, Tung, Nasrallah and Nasrallah2002; Shiba et al., Reference Shiba, Iwano, Entani, Ishimoto, Shimosato, Che, Satta, Ito, Takada, Watanabe, Isogai and Takayama2002).
The SSI reaction involves a kinase-dependent phosphorylation signalling cascade that is part of a ubiquitination-degradation type of cell–cell recognition system (Goring & Walker, Reference Goring and Walker2004). Control of SI depends on a complex interplay between promoters and inhibitors of the signal transduction pathway (Cabrillac et al., Reference Cabrillac, Cock, Dumas and Gaude2001; Takayama & Isogai, Reference Takayama and Isogai2003; Goring & Walker, Reference Goring and Walker2004; Murase et al., Reference Murase, Shiba, Iwano, Che, Watanabe and Isogai2004) that ultimately results in blocking of self-related pollen tube penetration by a process reminiscent of programmed cell death. Details of the downstream signalling pathway are reviewed elsewhere but the important factors to note are that there are many steps in the pathway where a disruption or mutation could result in loss of SI. It might therefore be expected that independent losses of SI could be achieved through more than one mechanism. Since most of the downstream genes are not located at the S-locus itself, changes in SRK and SCR might not be expected to be seen for some time after loss of SI has occurred if it is the signalling pathway that is disrupted.
Much of the original work on SI in the Brassicaceae was on cultivated plants in the genus Brassica (reviewed in Charlesworth & Awadalla, Reference Charlesworth and Awadalla1998). Although important clues to the biochemical control of SI in Brassica have come from studying self-compatible lines of the allotetraploid (amphidiploid) Brassica napus (reviewed in Brugière et al., Reference Brugière, Cui, Bi, Arnoldo, Jackman and Rothstein2000; Goring, Reference Goring2000), confounding influences of cultivation history and origins through hybridization make it difficult to interpret selection pressures contributing to the loss of SI. The comparable SI system has now been well-characterized in a naturally occurring plant, Arabidopsis lyrata (Charlesworth et al., Reference Charlesworth, Awadalla, Mable and Schierup2000; Kusaba et al., Reference Kusaba, Dwyer, Hendershot, Vrebalov, Nasrallah and Nasrallah2001; Schierup et al., Reference Schierup, Mable, Awadalla and Charlesworth2001; Nasrallah et al., Reference Nasrallah, Liu and Nasrallah2002; Charlesworth et al., Reference Charlesworth, Bartolomé, Schierup and Mable2003a, Reference Charlesworth, Mable, Schierup, Bartolomé and Awadallab; Mable et al., Reference Mable, Schierup and Charlesworth2003) and there has been increasing interest in extending this understanding to other closely related species (Castric & Vekemans, Reference Castric and Vekemans2004; Bechsgaard et al., Reference Bechsgaard, Castric, Charlesworth, Vekemans and Schierup2006; Paetsch et al., Reference Paetsch, Mayland-Quellhorst and Neuffer2006). A. lyrata has been assumed to be obligately outcrossing and, as one of the closest extant relatives of self-compatible A. thaliana, is increasing in popularity as a genetic model for comparative genomic approaches (Mitchell-Olds, Reference Mitchell-Olds2001; Kuittinen et al., Reference Kuittinen, de Haan, Vogl, Oikarinen, Leppala and Koch2004; Wright et al., Reference Wright, Yau, Looseley and Meyers2004; Koch & Kiefer, Reference Koch and Kiefer2005; Shimizu & Purugganan, Reference Shimizu and Purugganan2005; Yogeeswaran et al., Reference Yogeeswaran, Frary, York, Amenta, Lesser and Nasrallah2005; Bechsgaard et al., Reference Bechsgaard, Castric, Charlesworth, Vekemans and Schierup2006; Berr et al., Reference Berr, Pecinka, Meister, Kreth, Fuchs and Blattner2006). High inbreeding depression and high mortality of seeds arising from enforced self-pollinations have been found in European populations (A. lyrata subspecies petraea), supporting the idea that the consequences of losing SI would be harsh in species with a long history of outcrossing (Karkkainen et al., Reference Karkkainen, Kuittinen, van Treuren, Vogl, Oikarinen and Savolainen1999; Schierup et al., Reference Schierup, Mable, Awadalla and Charlesworth2001; Bechsgaard et al., Reference Bechsgaard, Castric, Charlesworth, Vekemans and Schierup2006). In contrast, naturally occurring populations of A. lyrata subspecies lyrata have been identified in the Great Lakes region of eastern North America that vary not only in strength of SI but in actual outcrossing rates in the field (Mable et al., Reference Mable, Robertson, Dart, Di Berardo and Witham2005; Mable & Adam, Reference Mable and Adam2007). There is thus the potential to use the Arabidopsis species group not only to compare the consequences of increased inbreeding between species but also within species occurring in the same geographic region.
Original theories about the dynamics of SI systems were based on Brassica but differences found in the SI systems of wild Arabidopsis species have already led to some theoretical revisions (Castric & Vekemans, Reference Castric and Vekemans2004). For example, in Brassica, haplotypes can be divided into a single predominantly dominant and a single predominantly recessive class whereas in A. lyrata there are at least two additional intermediate classes (Bechsgaard et al., Reference Bechsgaard, Bataillon and Schierup2004; Prigoda et al., Reference Prigoda, Nassuth and Mable2005; Schierup et al., Reference Schierup, Bechsgaard, Nielsen and Christiansen2006). This means that there are effectively more recessive combinations and more potential for low levels of inbreeding in the latter. The dynamics of allelic turnover and relative frequency of dominant and recessive alleles also could be quite different than if only two dominance classes were maintained. Based on data emerging from wild species, Billiard et al. (Reference Billiard, Castric and Vekemans2007) used simulations to investigate the dynamics of equilibrium frequencies of the number of alleles per dominance class. Their results suggest that higher numbers of alleles can be maintained in more dominance classes, that the number of dominance classes can evolve, and that recessive alleles can be at very high frequencies when there is a single most recessive class.
3. Mechanisms for loss of SI
If we understand more about what can go wrong in SI systems to render them non-functional, we are likely to gain insights into what is required to maintain them. For example, it has been observed that extremely low levels of variation in SRK sequences normally exist within the same haplotype sampled from different geographic regions, within or even between different outcrossing species, compared with large differences between haplotypes (Miege et al., Reference Miege, Ruffio-Chable, Schierup, Cabrillac, Dumas and Gaude2001; Kimura et al., Reference Kimura, Sato, Fujimoto and Nishio2002; Sato et al., Reference Sato, Nishio, Kimura, Kusaba, Suzuki and Hatakeyama2002). It is not known, however, whether this within-haplotype homogenization would persist (or for how long) after loss of SI and whether it is a cause or a consequence of preserving a functional SI system. In Brassica, relatively high variation appears to be tolerated within haplotypes for SCR sequences without changing their specificity to particular SRK sequences (Chookajorn et al., Reference Chookajorn, Kachroo, Ripoll, Clark and Nasrallah2004). This is in stark contrast to very low levels of polymorphism observed among SCR orthologues in A. thaliana. Although this has been used to suggest that a recent selective sweep of a non-functional SCR allele (ΨSCR1) subsequent to postglacial expansion could explain the complete loss of SI in A. thaliana (Shimizu et al., Reference Shimizu, Cork, Caicedo, Mays, Moore and Olsen2004), this has remained controversial (Charlesworth & Vekemans, Reference Charlesworth and Vekemans2005). Patterns of divergence between the three SRK haplotypes found in A. thaliana with orthologues in A. lyrata and A. halleri (termed ‘haplogroups’) are consistent with a selective sweep but could also be explained by mutations at another gene, with subsequent loss of diversity at the S-locus (Bechsgaard et al., Reference Bechsgaard, Castric, Charlesworth, Vekemans and Schierup2006). A survey of worldwide accessions of A. thaliana suggests that extensive remodelling of the S-locus has occurred since loss of SI but there does not appear to be recombination between ΨSCR1 and the three major haplogroups originally defined based on ΨSRK types (Sherman-Broyles et al., Reference Liu, Sherman-Broyles, Nasrallah and Nasrallah2007). These authors conclude that a selective sweep for a mutation at ΨSCR1 is less likely than changes at other genes, with subsequent reduction in diversity at ΨSCR.
A broader survey of A. thaliana accessions strongly supports this latter conclusion. Previous failure to identify variants at the S-genes has probably been due to high sequence divergence in some accessions and extensive rearrangements within the S-locus region (Tang et al., Reference Tang, Toomajian, Sherman-Broyles, Plagnol, Guo and Hu2007). The authors conclude that this rules out a selective sweep and favours instead gradual erosion of the ancestral balanced polymorphism that would have been maintained during the self-incompatible phase. They also examine genome-wide patterns of linkage disequilibrium (LD) and suggest that the decay in LD is more representative of a shift from outcrossing to selfing on the order of at least a million years. Since it has been estimated that SRK started to become a pseudogene within the past 400 000 years (Bechsgaard et al., Reference Bechsgaard, Castric, Charlesworth, Vekemans and Schierup2006), this would support loss of SI through a modifier rather than a mutation to render SRK non-functional. Given the diversity among A. thaliana accessions, it is also possible that SI was lost more than once and through different mechanisms, but this would make it even more difficult to disentangle cause from effect.
Loss of SI through the action of unlinked modifiers has been predicted theoretically (Uyenoyama, Reference Uyenoyama1991; Charlesworth et al., Reference Charlesworth, Morgan and Charlesworth1992; Levin, Reference Levin1996) and has received some empirical support. Nasrallah et al. (Reference Nasrallah, Liu, Sherman-Broyles, Boggs and Nasrallah2004) examined variation in response of self-compatible A. thaliana ecotypes to transformation with functional A. lyrata S-alleles (Nasrallah et al., Reference Nasrallah, Liu and Nasrallah2002) to assess the number of loci likely to be involved in the loss of SI. They found variation among ecotypes in the strength of SI following transformation and suggested that it was most likely that an external modifier was responsible for the shift to self-compatibility (Nasrallah et al., Reference Nasrallah, Liu, Sherman-Broyles, Boggs and Nasrallah2004). More recently, they have shown that this cryptic pseudo-self-compatibility is caused by a mutation in a gene (PUB8) encoding a U-box-containing protein that is linked to the S-genes and which regulates SRK transcript levels (Liu et al., Reference Liu, Sherman-Broyles, Nasrallah and Nasrallah2007). In addition, breakdown of SI in artificial interspecific crosses appears to be due to reversible epigenetic effects on transcript levels in either pollen or stigma S-genes (Nasrallah et al., Reference Nasrallah, Liu, Sherman-Broyles, Schmidt and Nasrallah2007). Modifiers, rather than mutations at the S-locus, have also been implicated in loss of SI for gametophytic systems (Good-Avila & Stephenson, Reference Good-Avila and Stephenson2002; O'Brien et al., Reference O'Brien, Kapfer, Major, Laurin, Bertrand and Kondo2002). However, one predominant mechanism for loss of SI has not yet been revealed in any system.
In A. lyrata lyrata, a survey of SRK alleles in three predominantly outcrossing and two predominantly selfing populations (Mable et al., Reference Mable, Robertson, Dart, Di Berardo and Witham2005) did not reveal differences in the number of dominant or recessive alleles between the two types of populations and there was a large degree of overlap in SRK alleles between populations. This again suggests that loss of SI is not due to mutations at SRK. Preliminary evidence investigating the inheritance of loss of SI through crosses between self-incompatible and self-compatible individuals confirms the lack of association with particular S-haplotypes but suggests that partial self-compatibility can obscure interpretation of what causes complete loss of SI. Even in strongly SI European populations (A. lyrata petraea), many individuals produce small fruits (with only a few seeds) that appear to represent leakiness rather than disruption of the SI system (Mable et al., Reference Mable, Schierup and Charlesworth2003). The cause of this is not known but it does not appear to be predominantly environmental, as the same individuals tend to produce small fruits whenever they are selfed. In addition, some individuals appear to show a weakening of the SI reaction, and show differences between replicates in whether or not a full-sized fruit with seeds is produced (partial compatibility, PC).
Crosses between individuals from a predominantly outcrossing population (PIN) with individuals from a predominantly selfing population (RON) compared with another predominantly outcrossing population (IND) highlight the complexity of patterns that emerge in the face of partial self-compatibility (Fig. 1; Table 1). Small fruits (indicated in parentheses) show limited viability of seeds; self-pollinations that resulted in small fruits were thus classified as self-incompatible (Mable et al., Reference Mable, Schierup and Charlesworth2003). In crosses involving RON individuals, progeny that were completely self-compatible were produced at high frequency regardless of whether the pollen donor was self-compatible or self-incompatible (Table 1, families 1–3). In crosses involving IND individuals (Table 1, families 4–6), on the other hand, except for a single self-compatible individual, progeny tended to be either self-incompatible or partially compatible, even when the pollen donor was capable of producing viable selfed seeds (Table 1, family 5). Except for a few cases, there was little correspondence between mean selfed fruit lengths in progeny compared with their mid-parental values. There was also no relationship between progeny phenotype and genotype at the SRK gene, and segregation ratios of parental genotypes did not deviate from expected (Table 2).

Fig. 1. Preliminary survey of distribution of cpDNA haplotypes in populations of A. lyrata lyrata sampled from the Great Lakes Region of eastern North America. Predominantly selfing populations are indicated by open circles; predominantly outcrossing populations are indicated by filled circles; mixed populations are indicated by hatched circles. Three haplotypes have been found in the region (indicated by numerals inside circles). All the predominantly selfing populations sampled so far have haplotype 2, along with the more southerly distributed predominantly outcrossing populations. The TSS population has a mixture of self-incompatible and self-compatible individuals, with the former having haplotype 1 and the latter having haplotype 2. A low frequency of self-compatible individuals also occurs in the LSP and MAN populations, which are otherwise predominantly outcrossing. See Mable & Adam (Reference Mable and Adam2007) for a complete description of localities.
Table 1. Segregation of SI phenotypes for crosses showing number of progeny compared (N); maternal and paternal SI phenotypes (P); viability of selfed seeds (V) produced by maternal and paternal parents; germination rates (% G) and survivorship (% S) for seeds from crosses; percentage of progeny that were classified as SI, SC and PCFootnote b; mean fruit length of selfed seeds of progeny; and mid-parent values for mean fruit length of seeds produced by selfing

a Plants labelled PIN and IND are from predominantly outcrossing populations; those labelled RON are from a predominantly selfing population.
b Number of seed-containing fruits produced on self-pollination, with the number classified as ‘small’ (having 3 or fewer seeds) in parentheses.
c SI classification based on Mable et al. (Reference Mable, Robertson, Dart, Di Berardo and Witham2005), where small fruits are considered to be negative: SI, self-incompatible (0 or 1 selfed fruits produced out of 6 replicates); SC, self-compatible (5 or 6 selfed fruits produced out of 6 replicates); PC, partially compatible (2–4 selfed fruits produced out of 6 replicates).
Table 2. Segregation of SRK genotypes in crosses listed in Table 1, showing parental genotypes, observed genotype combinations within families, the number of individuals (N) in which particular genotypes were seen, and the frequency of SI phenotypes within genotypic classes
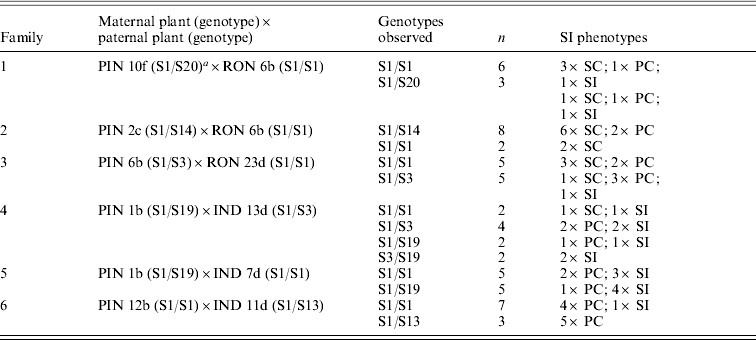
a This plant also had the S13 allele, which was transmitted to all progeny and appears to represent a duplication of SRK or of the S-locus.
Although these data support a modifier hypothesis more than changes at the S-locus, evaluation of crosses without partial self-compatibility are required to determine whether partial SI is an intermediate step on the road to complete self-compatibility or whether it could provide a mechanism to allow more mating partners in small populations. In addition, preliminary data based on chloroplast DNA (see below) suggest that not all populations are genetically equivalent, and so there is the potential that SI has been lost independently in different geographic regions.
4. Changes in diversity resulting from loss of SI
There has been increasing interest in comparing relative diversity and population structure in selfing A. thaliana with that in its outcrossing relatives (Savolainen et al., Reference Savolainen, Langley, Lazzaro and Fréville2000; Clauss et al., Reference Clauss, Cobban and Mitchell-Olds2002; Wright et al., Reference Wright, Lauga and Charlesworth2002; Wright et al., Reference Wright, Yau, Looseley and Meyers2004; Balaña-Alcaide et al., Reference Balaña-Alcaide, Ramos-Onsins, Boone and Aguadé2006; Bechsgaard et al., Reference Bechsgaard, Castric, Charlesworth, Vekemans and Schierup2006; Kawabe et al., Reference Kawabe, Hansson, Hagenblad, Forrest and Charlesworth2006; Wright et al., Reference Wright, Iorgovan, Misra and Mokhtari2007). Tests of selection and recombination can be powerful when using polymorphisms within species compared with divergence between species (McDonald & Kreitman, Reference McDonald and Kreitman1991), but results could be easier to interpret if such tests are conducted between populations of the same species. For example, Wright et al. (Reference Wright, Lauga and Charlesworth2002) evaluated the consequences of mating system differences in self-compatible A. thaliana compared with self-incompatible A. lyrata populations by comparing molecular evolution across a range of ‘neutral’ loci spanning a large portion of the genome. Contrary to expectations, they found no significant difference in the rates of protein evolution observed between selfing and outcrossing Arabidopsis species. However, they concluded that differences in demographic history may have obscured relationships when comparing A. thaliana ecotypes with samples of A. lyrata collected from European and North American populations, each of which may have experienced different types of local selection pressures (Wright et al., Reference Wright, Lauga and Charlesworth2003).
Many of the early comparisons on genetic diversity in selfing and outcrossing species in the Brassicaceae focused on the genus Leavenworthia, where it has been consistently demonstrated that selfing species show lower levels of diversity and heterozygosity than their outcrossing relatives (Charlesworth & Yang, Reference Charlesworth and Yang1998; Liu et al., Reference Liu, Zhang and Charlesworth1998; Filatov & Charlesworth, Reference Filatov and Charlesworth1999; Liu et al., Reference Liu, Charlesworth and Kreitman1999). Since orthologues of the SRK locus have recently been identified in this genus (J. Busch and D. Schoen, personal communication) and since there are multiple species with different outcrossing rates, this should continue to provide another powerful model system to understand the genetic consequences of loss of SI in relation to demographics and other traits that differ between species.
Comparisons between multiple populations of A. lyrata lyrata that differ in mating system are so far based only on microsatellite data (Mable et al., Reference Mable, Robertson, Dart, Di Berardo and Witham2005; Mable & Adam, Reference Mable and Adam2007) but they do suggest that it is important to disentangle effects due to demographic and life history differences, even within species. In the Great Lakes region of Eastern North America, A. lyrata lyrata occurs in geologically young habitats (i.e. 2000–10 000 years) that have been fragmented by both human expansion (sand dunes, rocky outcrops, alvars) and glacial retreat, so it is likely that it has experienced changes in population size and/or connectivity throughout its history. Multilocus microsatellite variation in progeny arrays has demonstrated that not only do some populations show a high frequency of individuals capable of selfing but realized outcrossing rates (t m) in the field are also well below 50% (Mable et al., Reference Mable, Robertson, Dart, Di Berardo and Witham2005; Mable & Adam, Reference Mable and Adam2007). There appear to be two distinctive types of plants in the area: (1) outcrossing plants (t m >0·8) with functional SI systems, relatively high diversity and requiring pollinators to set seed; and (2) inbreeding plants (t m <0·3) that have lost SI, show depressed genetic diversity and heterozygosity, and can set seed without pollinators (autogamous fruit set). The reduction in genetic diversity and heterozygosity in populations containing large numbers of the latter type of individuals (Mable & Adam, Reference Mable and Adam2007) is less than half that in predominantly outcrossing populations (ratio of He=0·3; Ho=0·17). Selfing populations also show higher degrees of differentiation from one another than they do from outcrossing populations or than outcrossing populations do from one another. Overall, microsatellite patterns suggest little gene flow between any populations except for two geographically close self-compatible populations (RON and PTP on Lake Erie; Fig. 1).
Although these patterns support theoretical predictions (Charlesworth & Wright, Reference Charlesworth and Wright2001), estimates of population structure based on microsatellite data could be confounded by large differences in number of genotypes in selfing compared with outcrossing populations, extensive differentiation of all populations from one another, and violation of random mating assumptions in selfing populations (Mable & Adam Reference Mable and Adam2007). Microsatellite data did not show evidence of recent bottlenecks in selfing populations (Mable et al., Reference Mable, Robertson, Dart, Di Berardo and Witham2005; Mable & Adam Reference Mable and Adam2007) but nucleotide data should provide more powerful demographic tests. Comparison of nucleotide diversity between one of the same selfing populations (RON) and one of the same outcrossing populations (IND) did not find a significantly lower nucleotide diversity in the predominantly selfing population (Wright et al., Reference Wright, Foxe, DeRose-Wilson, Kawabe, Looseley and Gaut2006). This could reflect a difference in time scales of nucleotide variation compared with microsatellites.
Preliminary data on the distribution of selfing populations in relation to chloroplast DNA variation (Fig. 1; Hoebe & Mable, unpublished data) suggests that there is a geographic component to genetic heterogeneity in the region. This means that interpretations of effects due to loss of SI must consider which outcrossing populations most likely gave rise to the current selfing populations. In a preliminary screen using primers for the Trn E-F region of the chloroplast DNA (Koch & Kiefer, Reference Koch and Kiefer2006), three distinctive haplotypes have been found in the 11 populations screened so far. The distribution of haplotypes is consistent with multiple colonization events into the region from different source populations, as has been found for a number of animal species (Austin et al., Reference Austin, Lougheed, Neidrauer, Chek and Boag2002). All of the predominantly selfing populations share the same haplotype (haplotype 2) as that found in more southerly distributed outcrossing populations. Interestingly, a population with mixed mating also shows a mixture of two haplotypes, with self-compatible individuals showing haplotype 2 and self-incompatible individuals showing haplotype 1 (Hoebe & Mable, unpublished data). In addition, some loss of SI appears to have occurred in LSP and MAN populations, which could represent independent events, since they do not share the same cpDNA haplotype. Further investigation of mechanisms for loss across the region could reveal whether there are common mechanisms or whether SI can be lost in more than one way.
The data are not yet sufficient to evaluate whether loss of SI occurred before or after postglacial expansion, but sampling more populations to the south of the Great Lakes and sequencing more gene regions will help to set the biogeographical framework necessary to establish how long ago and how many times SI was lost in the region. Sequence data based on emerging genomic information should allow use of more powerful tests to infer historical changes in population sizes and estimates of gene flow based on coalescent models that will provide more robust evaluation of population structure, changes in effective population size and genetic diversity in relation to mating system.
5. Inbreeding depression and loss of SI
The lack of robust conclusions about whether purging can be observed in natural populations (reviewed in Keller & Waller, Reference Keller and Waller2002; Goodwillie et al., Reference Goodwillie, Kalisz and Eckert2005) may not be surprising in the face of all the factors that can affect the rate and magnitude of expression of genetic load. Genetic load causing inbreeding depression can be difficult to purge if the number of recessive lethals accumulated during the SI phase is high. The relatively high frequency of loss of SI across species suggests that populations that survive a shift to self-compatibility should show at least some recovery from the high levels of inbreeding depression often observed with strong SI systems. Findings that pollen competition (Lankinen & Armbruster, Reference Lankinen and Armbruster2007), fluctuation in the strength of inbreeding between years in the same populations (Lyons & Antonovics, Reference Lyons and Antonovics1991) and differential consequences of inbreeding depression depending on the developmental stage considered (Husband & Schemske, Reference Husband and Schemske1996) all emphasize that purging might not be the only way to mitigate effects of inbreeding. Inbreeding depression can also be strongly influenced by maternal effects, which can be difficult to separate from differences due to population history (Picó et al., Reference Picó, Ouborg and van Groenendael2003).
The dynamics of inbreeding depression are often considered in relation to the population history of outcrossing, but it could be the outcrossing history of individuals that is most important. Although this idea has been promoted in a number of theoretical models (reviewed in Schultz & Willis, Reference Schultz and Willis1995), a simulation study by Schultz & Willis (Reference Schultz and Willis1995) suggested that variation in random genetic effects between individuals could be the primary cause of variation in inbreeding depression and that this variation can be higher in completely selfing or completely outcrossing populations compared with those with intermediate selfing rates. Empirical support for the primary forces driving variation in inbreeding depression among individuals is not yet sufficient to determine which of these scenarios is most representative, but both emphasize the importance of considering individual rather than population variation.
The most thorough empirical data on inbreeding depression in the Brassicaceae comes from work in the genus Leavenworthia. This work also emphasizes that differences in inbreeding depression can exist even between closely related species. Two species with high levels of natural self-fertilization (L. uniflora and L. crassa) retained substantial genetic load, as selfed progeny had lowered survival and fertility than outcrossed progeny in both species (Charlesworth et al., Reference Charlesworth, Lyons and Litchfield1994). In contrast, comparisons between five self-incompatible and five self-compatible populations of L. alabamica suggested that some purging of the genetic load has occurred with the shift in breeding system (Busch, Reference Busch2005a). Compared with outcrosses, selfed progeny from individuals from self-incompatible populations produced fewer and smaller seeds and showed significant inbreeding depression in both early developmental and adult fitness traits. Individuals from self-compatible populations, on the other hand, showed no differences in the size or number of seeds produced by outcrossing or selfing but showed marginally significant inbreeding depression in flower number caused by enforced self-fertilization. Since self-compatible populations have also been found to produce more fruit and seeds than self-incompatible populations (Busch, Reference Busch2005b), selfing populations in this species should receive all the benefits of reproductive assurance without major fitness costs. Research on A. lyrata has demonstrated that not all S-haplotypes are equal in terms of transmission and viability (Bechsgaard et al., Reference Bechsgaard, Bataillon and Schierup2004) and so it could also be important to consider whether inbreeding depression varies based on particular S-haplotypes. Unfortunately, the S-locus has not yet been characterized well enough in species of Leavenworthia to evaluate S-haplotype specific effects.
Understanding of the genetic causes of inbreeding depression and of which types of genes might be subject to purging following a shift from outcrossing to selfing could be increased by genome-wide scans of differences between selfed and outcrossed progeny of individuals that vary in outcrossing history. A very preliminary study using three A. lyrata lyrata mothers capable of producing viable selfed fruits (two from predominantly outcrossing populations and one from a predominantly selfing population) found no significant differences in germination rates (Fig. 2a), seedling weights or microarray-based transcriptome profiles (AtH1 gene chips; Affymetrix) between selfed and outcrossed progeny (Mable, unpublished data). While this is consistent with the L. alabamica work and could suggest purging, these data highlight the importance of considering parental effects, including inbreeding history and geographic locality. Both LSP and TSS populations are in close proximity (within 2–10 km) to populations showing a mixture of self-incompatible and self-compatible individuals but are themselves predominantly outcrossing; the RON population is composed of predominantly self-compatible individuals and is highly inbreeding, as are all of its neighbouring populations. Purging of genetic load might thus be expected to be seen more in the latter population and would be reflected in fewer differences between selfed and outcrossed progeny. There were significant differences between mothers in the length of time over which seeds germinated, with the northern LSP population showing a significantly shorter germination period than the TSS and RON populations (Fig. 2d). At the time of this study, the cpDNA differences between populations had not yet been uncovered, so differences in genetic background between populations could have obscured differences due to mating system history (see Fig. 1).

Fig. 2. Differences in percentage germination (a) and length of germination period (b) between selfed and outcrossed progeny for three mothers capable of setting seed from one predominantly outcrossing population (LSP), one mixed population (TSS) and one predominantly inbreeding population (RON), corresponding to the labels on Fig. 1. There were no significant differences between cross types for fitness measures, or for percentage germination between mothers (c). There was a significant difference between mothers in the length of the germination period (d), with LSP showing significantly faster germination than the others.
In the microarray pilot study, hybridization of genomic DNA, to account for differences in hybridization of the A. lyrata genome to the A. thaliana gene chips, also showed differences between mothers. Offspring from each mother did show a limited number of genes that varied in expression between selfed and outcrossed progeny, but there were no genes or pathways differing in expression that were shared between the progeny of all three mothers (Mable, unpublished data). There was a single gene (an HD-ZIP protein, IPR006712) that differed significantly between selfed and outcrossed progeny for both RON and LSP mothers, but this was not found for TSS. Whether or not microarrays are the best approach to uncovering genes affecting relative fitness, these results highlight that variation among populations or maternal effects could strongly influence interpretation of results.
6. Conclusions
Given the long history of theoretical interest in understanding the genetic consequences of changes in mating systems, we still know remarkably little about the range of mechanisms involved in loss of SI at the molecular level and specifically how factors such as population structure, phylogeography, demographic history and partial compatibility affect genetic diversity and fitness following a shift to inbreeding. However, recent research in the Brassicaceae suggests that we are currently on the brink of major breakthroughs in such understanding. This is at least partly due to advances in sequencing technology (which have allowed a broader genomic perspective on genetic consequences of mating system shifts) but also due to a shift in emphasis from cultivated plants to wild populations, where selection pressures have not been altered so dramatically by human activities. It is already apparent that individual variation such as maternal effects, background genetics, history of outcrossing and possibly particular S-genotypes will be important to consider, as well as variation between populations. Although it is not always easy to separate these effects, careful experiments using individuals from natural populations holds the most promise to understand the nature of the selective forces regulating mating system evolution.
The author thanks Andy Cossins and Margaret Hughes at the Liverpool Microarray node of the NERC Molecular Genetics facility for assistance and pump-priming funds for the microarray pilot project. The author also thanks Deborah Charlesworth for introduction to the field of plant population genetics and mentoring in the complex world of self-incompatibility. The comments of two anonymous reviewers significantly improved the manuscript. This work was supported by the Natural Environment Research Council.






