Published online by Cambridge University Press: 14 April 2009
Kimura used the heterozygosity and the number of low-frequency alleles to estimate that about 14% of mutations are selectively neutral. The method is shown to be subject to biases and to disruption due to bottleneck effects. Let deleterious alleles have selective disadvantage, s, compared with neutral alleles and let Ne denote the effective diploid population size. The estimator, 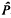 , of the proportion of neutral alleles is positively biased if (roughly) 4NeS < 25 or if 4Nes > 200. In the former case, one cannot adequately detect the different influences of deleterious and neutral alleles, whereas in the latter case, deleterious alleles will rarely appear in the sample. These difficulties cause the biases in
, of the proportion of neutral alleles is positively biased if (roughly) 4NeS < 25 or if 4Nes > 200. In the former case, one cannot adequately detect the different influences of deleterious and neutral alleles, whereas in the latter case, deleterious alleles will rarely appear in the sample. These difficulties cause the biases in 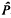 , and are likely to cause similar biases for any estimation method based solely on allele frequencies. There is substantial sampling variability in
, and are likely to cause similar biases for any estimation method based solely on allele frequencies. There is substantial sampling variability in 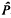 in cases of practical interest, when data from 11 loci, or even as many as 31 loci, are pooled. If there has been a recent contraction in population size,
in cases of practical interest, when data from 11 loci, or even as many as 31 loci, are pooled. If there has been a recent contraction in population size, 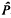 will be positively biased, often yielding values greater than 1 or even being infinite. But after a recent expansion in population size, the heterozygosity will not have made as quick an increase and
will be positively biased, often yielding values greater than 1 or even being infinite. But after a recent expansion in population size, the heterozygosity will not have made as quick an increase and 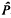 will be negatively biased. Population expansion alone can produce
will be negatively biased. Population expansion alone can produce 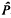 values close to those observed by Kimura, even if all alleles are neutral. In an appendix, a new method for simulating samples of neutral and deleterious genes is described.
values close to those observed by Kimura, even if all alleles are neutral. In an appendix, a new method for simulating samples of neutral and deleterious genes is described.