Introduction
RLS is characterized by a strong urge to move and uncomfortable sensations in lower limbs, which can be relieved by movements. Genome-wide association studies have implicated up to 19 risk loci for RLS, across which two of the candidate genes are MEIS1 and BTBD9 (Schormair et al., Reference Schormair, Zhao, Bell, Tilch, Salminen, Putz, Dauvilliers, Stefani, Högl, Poewe, Kemlink, Sonka, Bachmann, Paulus, Trenkwalder, Oertel, Hornyak, Teder-Laving, Metspalu and Winkelmann2017).
Knockout (KO) animals of BTBD9 or MEIS1 homologs exhibit RLS-like phenotypes. For instance, homozygous Btbd9 KO mice have motor restlessness, disrupted sleep, and altered sensory perception (DeAndrade et al., Reference DeAndrade, Johnson, Unger, Zhang, L., van Groen, Gamble and Li2012). Loss of BTBD9 homolog in Drosophila melanogaster results in increased motor activity, decreased dopamine levels, and disrupted sleep (Freeman et al., Reference Freeman, Pranski, Miller, Radmard, Bernhard, Jinnah, Betarbet, Rye and Sanyal2012). Heterozygous Meis1 KO mice are hyperactive (Meneely et al., Reference Meneely, Dinkins, Kassai, Lyu, Liu, Lin, Brewer, Li and Clemens2018). C. elegans with decreased MEIS1 homolog show increased expression of ferritin (Catoire et al., Reference Catoire, Dion, Xiong, Amari, Gaudet, Girard, Noreau, Gaspar, Turecki, Montplaisir, Parker and Rouleau2011). Therefore, both BTBD9 and MEIS1 may play a role in the development of RLS, yet whether and how the two genes interact is not known.
Objective
Our goal was to define the relationships between BTBD9 and MEIS1 in the pathogenesis of RLS. Egg retention assay in C. elegans was used to determine if there are genetic interactions between hpo-9, a BTBD9 homolog, and unc-62, a MEIS1 homolog. Furthermore, we created mouse models by knocking out BTBD9 homolog, Btbd9, MEIS1 homolog, Meis1, or both. Their motor-sensory responses were compared by wheel-running and tail-flick tests. Homozygous Meis1 KO was not included because of embryonic lethality (Spieler et al., Reference Spieler, Kaffe, Knauf, Bessa, Tena, Giesert, Schormair, Tilch, Lee, Horsch, Czamara, Karbalai, Toerne, Waldenberger, Gieger, Lichtner, Claussnitzer, Naumann, Müller-Myhsok and Winkelmann2014). The transcription of Btbd9 in Meis1 KO and the level of Meis1 protein in Btbd9 KO animals were studied.
Methods
C. elegans were maintained using standard methods (Catoire et al., Reference Catoire, Dion, Xiong, Amari, Gaudet, Girard, Noreau, Gaspar, Turecki, Montplaisir, Parker and Rouleau2011). The wildtype (WT) used was Bristol N2. The hpo-9 KO, hpo-9(tm3719), was obtained from the National BioResource Project (Japan) and backcrossed four times to the N2 background. RNAi against unc-62 (unc-62 RNAi) and the empty vector (EV) were used according to a standard feeding method with HT115 bacterial strain. Egg retention assay was performed according to published protocols (Chase & Koelle, Reference Chase and Koelle2004) and analyzed by a Students’ t-test (supplementary material).
Heterozygous Btbd9 KO (Lyu et al., Reference Lyu, Xing, DeAndrade, Liu, Perez, Yokoi, Febo, Walters and Li2019) were bred with Meis1 KO animals (Meneely et al., Reference Meneely, Dinkins, Kassai, Lyu, Liu, Lin, Brewer, Li and Clemens2018) to generate double heterozygotes, which were bred with heterozygous Btbd9 KO mice to generate experimental mice. Behavioral tests were conducted as described (Lyu et al., Reference Lyu, Xing, DeAndrade, Liu, Perez, Yokoi, Febo, Walters and Li2019) and analyzed by SAS GENMOD or mixed model ANOVA. Western blot and quantitative RT-PCR were performed and analyzed as described (Yokoi et al., Reference Yokoi, Dang, Liu, Gandre, Kwon, Yuen and Li2015) using striatal tissues (supplementary material).
Results
Worms: Figure 1 shows that unc-62 knockdown led to an increased number of eggs retained in both N2 and hpo-9(tm3719) as described (Kamath et al., Reference Kamath, Fraser, Dong, Poulin, Durbin, Gotta, Kanapin, Le Bot, Moreno, Sohrmann, Welchman, Zipperlen and Ahringer2003). The hpo-9 mutation caused fewer eggs retained in the presence or absence of unc-62 RNAi. Additionally, hpo-9(tm3719) treated with unc-62 RNAi retained a similar number of eggs as N2.
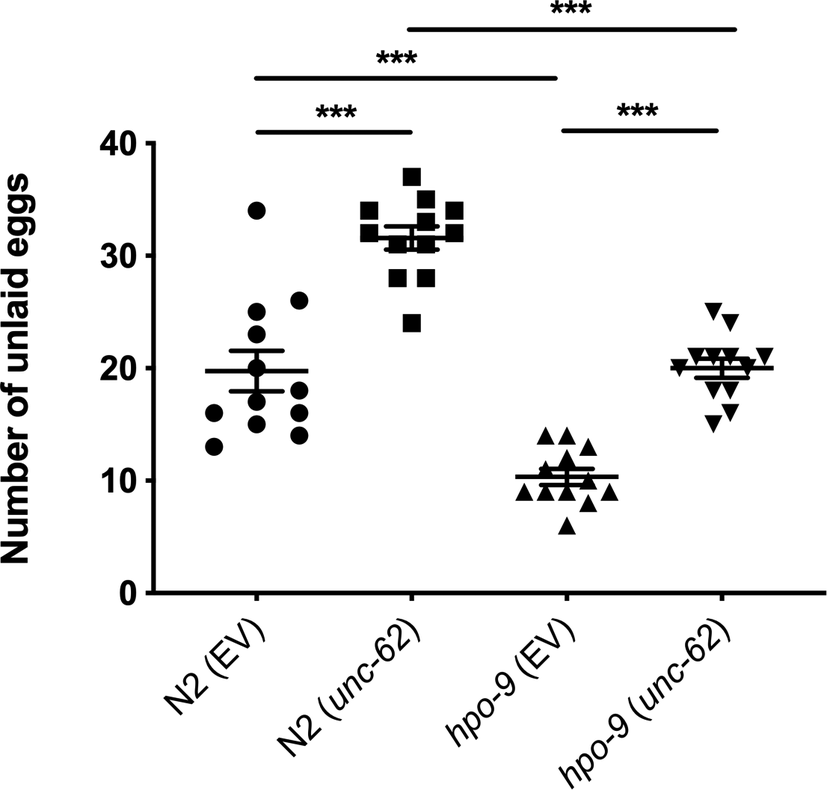
Figure 1. Egg retention assay. Bars represent the mean ± standard error of the mean (SEM) for 12 animals for each strain. ***, p < 0.001.
Mice: During the light phase of the wheel running test, neither single KOs exhibited significant difference compared with the WT (Figure 2). However, the double KO showed a robust increase from the WT and a lesser extent of increase from both single KOs. During the dark phase, activity levels were similar among the four groups. Figure 3 shows that the double KO did not have changes in the tail-flick response although both single KOs had reduced latency. Moreover, Meis1 protein levels and Btbd9 mRNA levels were unaffected by Btbd9 knockout and Meis1 deficiency, respectively (Figure 4).
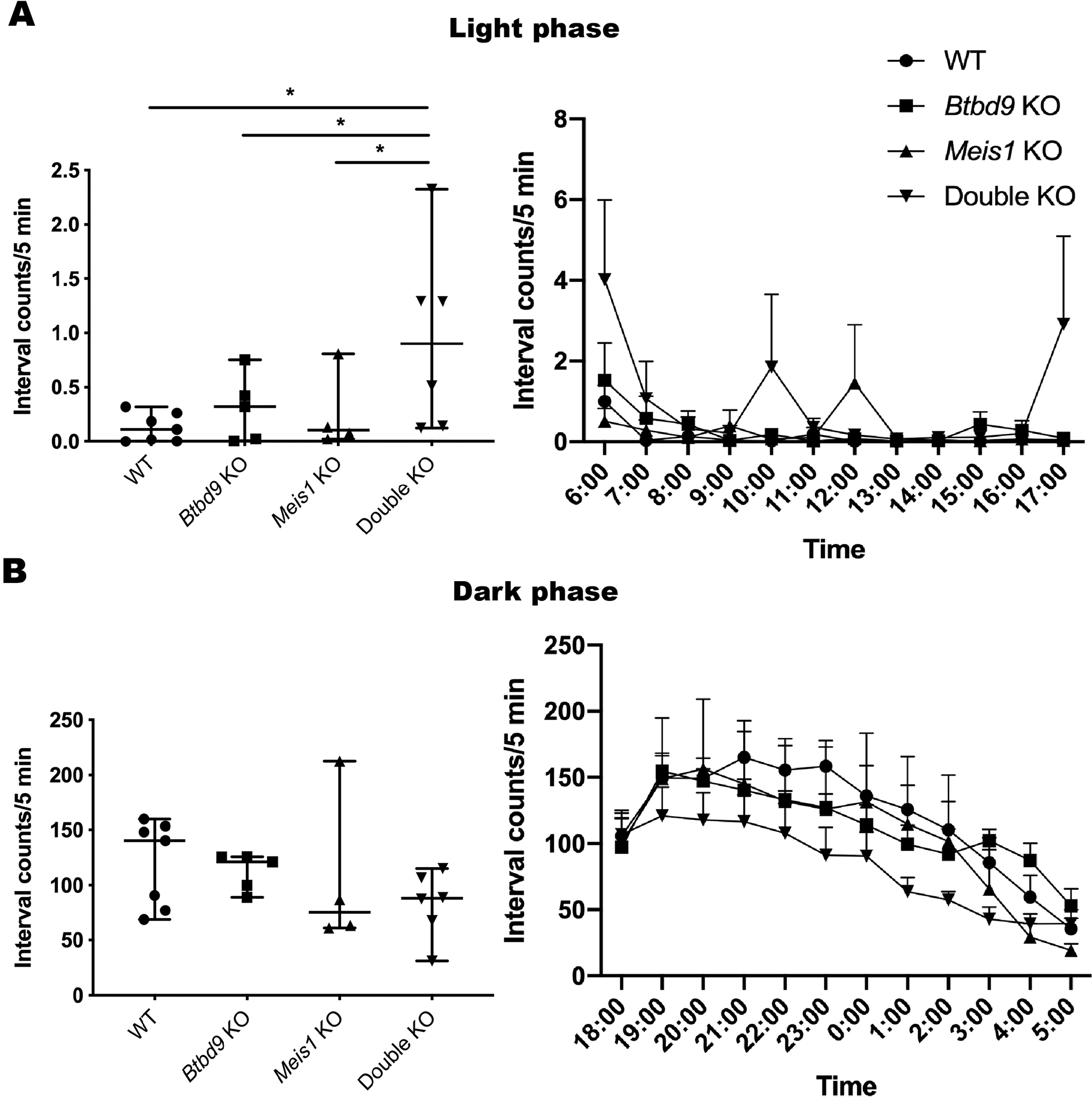
Figure 2. Wheel running during the light phase (A), and the dark phase (B). The data was not normally distributed and analyzed by SAS GENMOD with a negative binomial distribution. In the scatter plot, each dot is an average value calculated from 4 days’ data for each mouse. Bars represent the median with 95% confidence intervals (CIs). Hourly activity is presented next to the scatter plot. Each dot is an average value calculated from 4 days’ data for each genotype. The activity of the double KO mice shot up right after the light was turned on and right before the light was turned off. In addition, they also showed high levels of activity around the middle of the rest period. The results indicate that the double KO mice may have difficulty in falling asleep and tend to wake up early. WT, n = 7; Btbd9 KO, n = 5; Meis1 KO, n = 4; double KO, n = 6. *, p < 0.05.
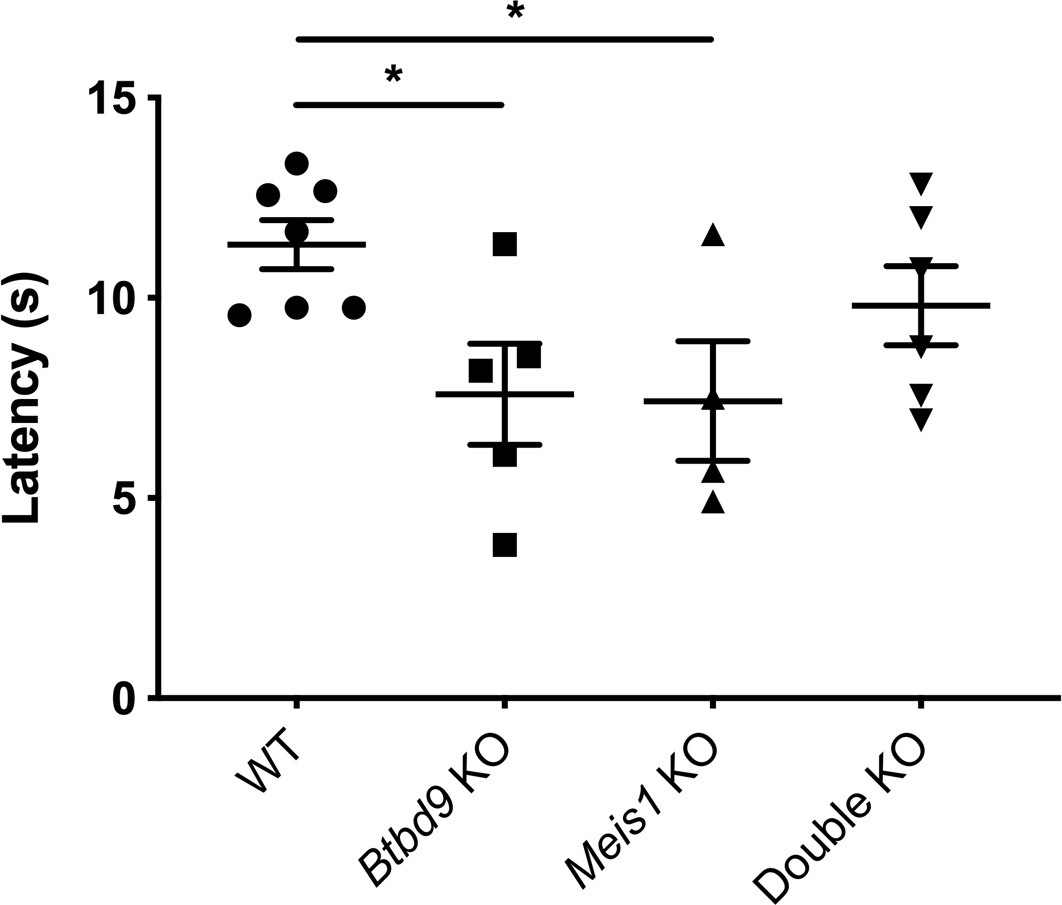
Figure 3. Tail-flick test. The data were normally distributed and analyzed by mixed model ANOVA with repeated measurements. Each dot is an average value calculated from 3 trials for each mouse. Single KO had reduced latency compared with the WT but did not show a significant difference compared with the double KOs. The double KO did not have significant changes compared with the WT. Bars represent the mean ± SEM. WT, n = 7; Btbd9 KO, n = 5; Meis1 KO, n = 4; double KO, n = 6. *, p < 0.05.
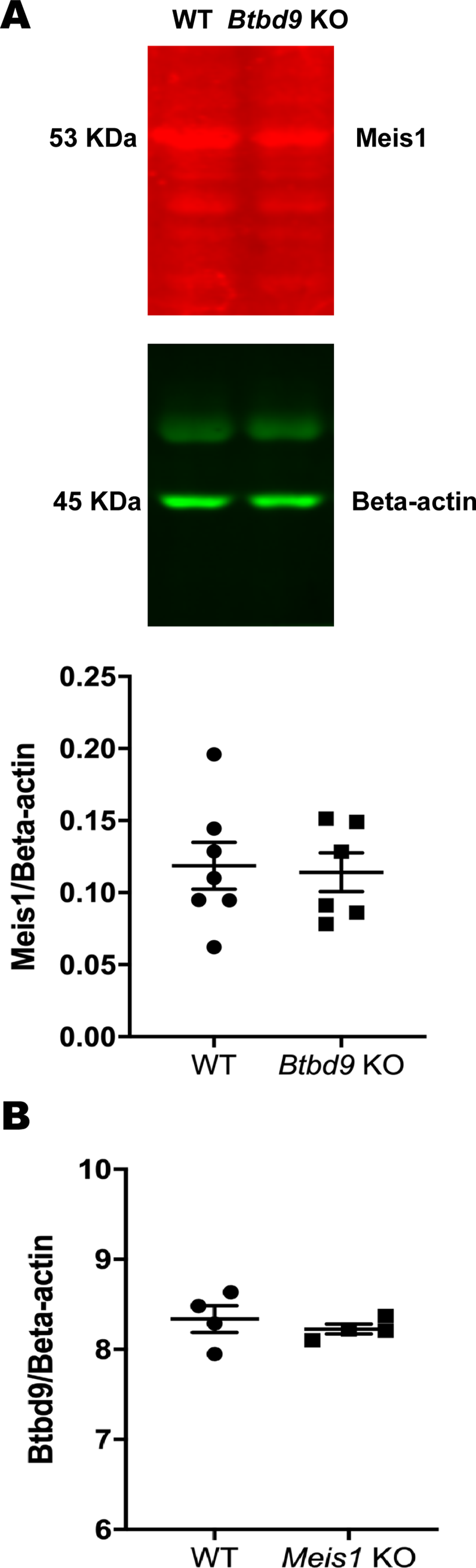
Figure 4. Molecular analysis. (A) Western blot to measure the amount of Meis1, normalized to β-actin, in Btbd9 KO (n = 6) and WT (n = 7) mice. (B) Quantitative RT-PCR to test the level of Btbd9 mRNA, normalized to β-actin, in Meis1 KO (n = 4) and WT (n = 4) mice. Bars represent the mean ± SEM.
Discussion
Wheel-running data from day and night were analyzed separately because RLS symptoms mostly happen at night, which is the day for rodents. With animals at an average age of 3 months, we did not observe increased activity in either single KO as suggested before (DeAndrade et al., Reference DeAndrade, Johnson, Unger, Zhang, L., van Groen, Gamble and Li2012; Meneely et al., Reference Meneely, Dinkins, Kassai, Lyu, Liu, Lin, Brewer, Li and Clemens2018), indicating that the double KO had early-onset deficit while the single KOs were still asymptomatic. It has been shown that Btbd9 expression does not change by Meis1 deficiency (Spieler et al., Reference Spieler, Kaffe, Knauf, Bessa, Tena, Giesert, Schormair, Tilch, Lee, Horsch, Czamara, Karbalai, Toerne, Waldenberger, Gieger, Lichtner, Claussnitzer, Naumann, Müller-Myhsok and Winkelmann2014). This was confirmed by molecular analyses, suggesting that Btbd9 and Meis1 do not regulate each other.
Conclusion
In worms, the augmentation effect of unc-62 knockdown is independent of hpo-9 manipulation and it is also true the other way around. Moreover, hpo-9 knockout and unc-62 knockdown counteract each other. In mice, the wheel running test suggests that there is an additive effect of Meis1 and Btbd9 mutations in the double KO mice. Btbd9 does not influence the Meis1 protein level, and Meis1 cannot alter Btbd9 gene expression. Hence, the two RLS risk genes work independently and have functional interactions in both worms and mice. Protein–protein interaction assays would be ideal to confirm this conclusion in the future.
Acknowledgments
We would like to thank Drs. Neil Copeland and Hesham Sadek for supplying Meis1 loxP mice, Dr. Shohei Mitani for the hpo-9(tm3719) strain, and Fumiaki Yokoi, Lin Zhang, Chad C. Cheetham, Sung Min Han, Jack Vibbert, Pauline Cottee, Jessica Winek, and Hieu Hoang for their technical assistance and stimulating discussions.
Author Contributions
SL, MPD, MAM, RX and YLi conceived and designed the study. SL, AD, YS, MPD, YY and YLiu conducted data gathering. SL and YLiu performed statistical analyses. SL and YLi wrote the article.
Funding Information
This work was supported by a grant from Restless Legs Syndrome Foundation; startup funds from the Departments of Neurology at UAB and UF; and the National Institutes of Health (grant numbers NS37409, NS47466, NS47692, NS54246, NS57098, NS65273, NS72872, NS74423, and NS82244). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Publishing Ethics
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.
Conflicts of Interest
All authors declare none.
Data Availability Statements
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Materials
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/exp.2020.12.






Comments
Comments to the Author: The manuscript details mouse and worm experiments on BTBD9 and MEIS1, two major candidate genes of RLS. As RLS is a polygenic disease, the double knock-out experiment is of great value in determining the relationship as well as a possible additive effect of these genes to the susceptibility to RLS. However, there are concerns relating to the analysis of the data which should be addressed.
In Figures 2 and 3, individual measurements resulting from repeated measurements in the same animal should not be displayed as individual data points, but should be first averaged within animal.
In statistical tests relating to Figs 2-3, was repeated measures ANOVA used? The test used should be stated in the legend. If so, the appropriate way to analyse the data would be to average first within animal and then use for example one-way ANOVA.
Division into light and dark period is used for wheel running activity, as is often done for this kind of data. Rather, the period right before and after the lights-on should be looked at. This would correspond better to human RLS and allow comparison with previous data in MEIS1 knock-out mice.
In the introduction, the sentence describing the human genetics of RLS is somewhat confusing and should be reformulated.
What does “4 repeats for each genotype” mean? Does this mean the last 4 days that were used in the analysis according to Lyu et al. 2019 or did the mice spend four times seven days in the running wheel?