INTRODUCTION
Anaplasma phagocytophilum is a widespread tick-borne pathogen reported to infect various species of wild and domestic animals [Reference Woldehiwet1]. It is a Gram-negative, obligate intracellular bacterium of the order Rickettsiales that infects white blood cells (neutrophils, eosinophils, monocytes). The primary vectors are ticks including Ixodes ricinus (Europe [Reference Woldehiwet1]), Ixodes scapularis (eastern United States), and Ixodes pacificus (western United States) [Reference Teglas and Foley2]. In cattle and sheep, A. phagocytophilum infection can cause tick-borne fever (TBF), a disease characterized by fever, depression, lethargy, reduced milk yield (dairy cattle) and abortion [Reference Woldehiwet1]. In horses, infection with A. phagocytophilum (earlier known as Ehrlichia equi) may cause equine granulocytic ehrlichiosis [Reference Gribble3], and in humans it is a cause of human granulocytic anaplasmosis (HGA [Reference Chen4]). HGA is considered to be an emerging vector-borne infection in Europe [Reference Dumler5] and the USA [Reference Dumler5, Reference Thomas, Dumler and Carlyon6], and is reportedly under-diagnosed due to the similarity of its clinical symptoms with infection with Borrelia burgdorferi [Reference Thomas, Dumler and Carlyon6].
The epidemiology of A. phagocytophilum in wildlife is poorly understood, and few reports on the pathogenic potential of A. phagocytophilum infection in cervids exist. These include case reports of A. phagocytophilum-associated mortality in roe deer (Capreolus capreolus) fawns and a moose calf (Alces alces) from Norway [Reference Jenkins7–Reference Stuen9]. In these reports, the animals succumbed due to a fatal, presumably secondary bacterial pneumonia, and the authors suggested that the immunosuppressive effects of a primary infection with A. phagocytophilum may have facilitated the lung infection. Seroprevalence in moose was reported to be 43% in a study in Norway [Reference Stuen10], but association with disease was not investigated. Another Norwegian study reported a 79% seroprevalence in moose from a habitat know to be tick-infested [Reference Milner and van Beest11]. Different genetic variants of A. phagocytophilum have been found in cervids based on 16S rRNA gene sequencing [Reference Petrovec12–Reference Welc-Falęciak14]. Red deer (Cervus elaphus) in the Czech Republic reportedly carry two different 16S rRNA variants of A. phagocytophilum [Reference Zeman and Pecha15]. However, sequences of other less conserved genes such as msp4, groESL or anka from different A. phagocytophilum isolates revealed substantial variability [Reference Ladbury16–Reference Stuen19]. For instance, up to 11 different variants of A. phagocytophilum based on the groESL gene sequences were detected in roe deer, red deer, alpine chamois (Rupicapra rupicapra), Alpine ibex (Capra ibex), and European mouflon (Ovis musimon) in Austria [Reference Silaghi13].
In 2005, a dead moose calf from the island of Öland in the Baltic Sea (Fig. 1) was submitted for post-mortem examination to the National Veterinary Institute in Uppsala, Sweden. The cause of death was a severe bacterial bronchopneumonia, with a concurrent septicaemia (caused by Streptococcus sp.). A. phagocytophilum DNA was detected by polymerase chain reaction (PCR) analysis [Reference Bernodt20]. It was suggested that the A. phagocytophilum infection facilitated the bacterial bronchopneumonia, resembling the findings of the report from Norway [Reference Jenkins7]. On the island of Öland local hunters have since 2006 reported low numbers of observed moose calves during the autumn hunt [21]. Several hunters on the island are also farmers (sheep, cattle), and frequently observe and treat their animals for tick-borne fever; similar information has been reported by local veterinarians [22]. Hence, it was suggested that infection with A. phagocytophilum had affected moose calf health and survival on the island.

Fig. 1. Map showing sampling areas (highlighted) in Sweden. A represents the island of Öland; B, C, and D represent sampling areas in three different mainland populations.
The epidemiology of A. phagocytophilum infection in moose populations based on both serological and PCR-based data has not been reported previously. The aim of the present study was to investigate the serological and DNA-based prevalence of A. phagocytophilum in hunter-harvested moose from the island of Öland, and three areas on the mainland of Sweden.
MATERIALS AND METHODS
Samples from harvested moose were collected from the island of Öland (Fig. 1, area A; 2007–2010), and from three mainland populations (Fig. 1, areas B, C, D; 2008–2010) during the moose hunt in October and November. Area A, the island of Öland, is a habitat with a mix of agricultural land and deciduous forest in the southern part, and boreal forest in the northern parts. Areas B, C, and D consist of mostly boreal forest [Reference Nilsson and Cory23]. Trained field assistants collected fresh samples (spleen, kidney, liver, faeces, lower jaws, ectoparasites, rumen contents, blood) from harvested moose, according to a standardized protocol. Blood (primarily from vena cava caudalis or the heart) was collected in non-additive blood tubes (BD Vacutainer®, BD Diagnostics, USA). Serum was separated from the blood samples and stored at −20°C. A 4 × 4 cm piece of the spleen, mandibles, and remaining samples were stored at −20°C awaiting further analysis. The body condition (below normal or normal) based on the presence of coronary fat deposits and signs of muscular atrophy, was recorded, and a specific identity was assigned to each sampled moose. The hunters recorded the weight of the carcass after removal of skin, head, internal organs, and metapodials, according to Wallin [Reference Wallin24].
For age determination in adult moose, the first molar of the lower jaw was sectioned and cementum layers counted as described by Wolfe [Reference Wolfe25]. The animals were divided into three age groups: calf (<1 year), subadult (1–2 years), and adult (>2 years).
The screening for presence of A. phagocytophilum DNA was performed with a diagnostic commercial real-time PCR test for detection of 16S rDNA using GER3 (TAGATCCTTCTTAACGGAAGGGCG) and GER4 (AAGTGCCCGGCTTAACCCGCTGGC) primers as described by Goodman et al. [Reference Goodman26], but converted to a real-time PCR format by adding a TaqMan probe [Reference Jäderlund27]. Total DNA was extracted from spleen samples, excising a cube with sides of 0·5 cm, mashing the tissue with sterile scalpels, subsequently followed by lysis and DNA extraction according to the manufacturer's instructions, using the QIAamp DNA Extraction kit and an EZ1 BioRobot (Qiagen GmbH, Germany). PCR was performed according to Jäderlund et al. [Reference Jäderlund27] in an ABI 7500 FAST PCR machine (Life Technologies, USA), and was run for 40 cycles. Each PCR included a control with the DNA template substituted with sterile water, a positive and a negative reference A. phagocytophilum template, extracted from blood from A. phagocytophilum-infected and non-infected horses, respectively. To prevent contamination from environmental DNA, the preparation of PCR mixtures, DNA extraction, PCR and detection of products were all performed in separated laboratory facilities, and aerosol-resistant filter pipette tips were used throughout the experiment.
Of the 10 spleen samples submitted for genetic sequencing, direct sequencing using GER3 and GER4 primers for 16S rDNA, yielded good quality sequences without the need for cloning for two samples (915 bp), one from a mainland moose and one from an island moose. From these two samples also partial groESL sequences were obatined (1275 bp). The PCR and sequencing protocol used was identical to that used by Franzén et al. [Reference Franzén28].
For detection of Anaplasma antibodies, an immunofluorescence assay (IFA) was used [Reference Jäderlund27]. Briefly, twofold dilutions of sera were added to slides precoated with A. phagocytophilum antigen (Protatek, USA). To visualize the bound antibodies, a fluorescein isothiocyanate (FITC)-conjugated rabbit anti-deer IgG (KP Laboratories, USA) was added and slides were examined by fluorescence microscopy. For negative control, a seronegative moose sample was used, and for a positive control the opposite. All samples were screened at a dilution of 1:40. For further titration, 77 high-quality (minimal hemolysis) serum samples from 2009 and 2010 were selected (51 from area A, 26 from areas B, C, D). These samples were further titrated in twofold steps to a dilution of 1:1280. An IFA titre ⩾1:40 was considered positive, in accordance with Stuen et al. [Reference Stuen10]. A further titration of samples with titres 1:1280 was not performed.
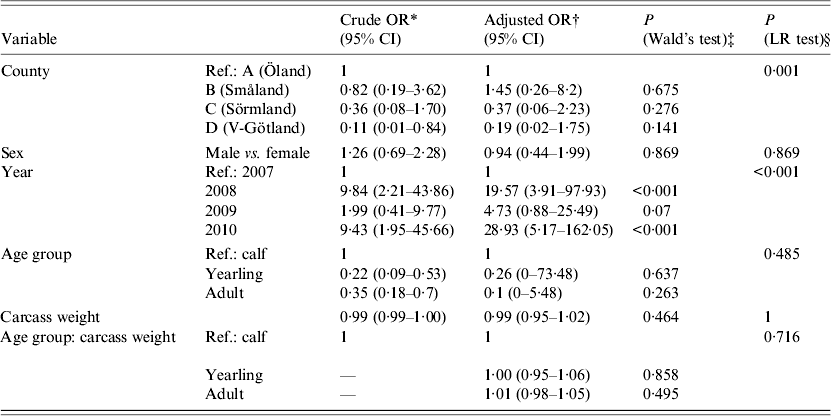
Statistical analyses of risk factors affecting PCR presence of A. phagocytophilum were performed using logistic regression in R (R Foundation for Statistical Computing, Austria). An exploratory analysis using multiple possible risk factors was conducted (Table 1). Crude and adjusted odds ratios (OR) with 95% confidence intervals were calculated to identify which factors (sampling area, sex, sampling year, age group, carcass weight) were associated with higher or lower PCR presence of A. phagocytophilum. The crude OR is an estimation of one specific factor without consideration and possible confounding effects of other factors. The adjusted OR is estimated with consideration of other factors (stated previously) included in the model. In addition, statistical significance calculations (P values) between sampling area, sex, sampling year, age group, and carcass weight (in relation to age group) were performed using Wald's test within the same logistic regression, as seen in Table 1.
Table 1. Results of a logistical regression model with factors associated with PCR-based detection of Anaplasma phagocytophilum in Swedish moose (Alces alces) spleens
OR, Odds ratio; CI, confidence interval; LR, likelihood ratio.
* The OR of one specific factor without consideration of confounding effects of other factors.
† The OR of one factor, with consideration of confounding effects of other factors.
‡ The P value based on the sample estimate.
§ Likelihood ratio test.
RESULTS
Spleen samples from 263 harvested moose (141 females, 122 males) were collected (Table 2). Blood was obtained from a total of 234 moose. From the remaining 29 moose, blood could not be collected, mostly due to contamination by rumen contents, or an insufficient collectable volume that occurred when the moose had been eviscerated prior to sampling by field assistants. Mean carcass weight for calves (n = 73) was 60·5 kg (range 25–87 kg), for sub-adults (n = 57) 127·3 kg (range 75–180 kg), and for adults (n = 109) 193·1 kg (range 118–302 kg). For 24 moose, no carcass weight was reported by the hunters. Twelve moose were found to be in subnormal body condition, and of these, clinical signs of disease (emaciation of unknown cause) were observed in two moose. The mean age of sampled moose was 2·8 years (range 0·5–15·5).
Table 2. Geographical and biological distribution, and numbers of Anaplasma phagocytophilum DNA-positive samples in moose spleen samples from Sweden collected from 2007 to 2010
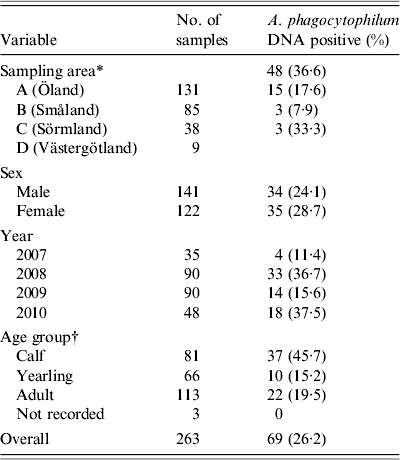
* Four different counties in southern Sweden according to Figure 1.
† Sampled moose divided in to three age groups: 0–1 year, 1–2 years, and >2 years.
The overall prevalence of A. phagocytophilum DNA was 26·3%. On a spatial scale, moose from area A (island) had a significantly (P < 0·006) higher prevalence of A. phagocytophilum DNA (36·6%) than the combined prevalence in moose from areas B, C, and D (15·9%). The area variable was highly significant (likelihood ratio test P < 0·01), indicating significant spatial heterogenity of A. phagocytophilum prevalence. Area A (island) had the highest recorded prevalence, 36·6%, and area C had the lowest recorded prevalence of 7·9%. No significant difference in prevalence (P = 0·141–0·657, Table 1) between individual counties was found. Regardless of sampling areas, samples from 2008 and 2010 had significantly (P < 0·01) higher A. phagocytophilum DNA prevalence than samples collected in 2007 (Table 1) and 2009 (results from separate model using 2009 as reference, not shown). For all moose samples from 2007 to 2010, sex, age group, and age did not have significant effects on the presence of A. phagocytophilum DNA.
All tested samples were serologically positive (titre ⩾1:40). Titration was performed on 77 samples from 2009 and 2010, with titres ranging from 1:40 to 1:1280. Figure 2 shows an overview of serological titres related to presence of A. phagocytophilum DNA. An additional model fitted to a subset of 77 animals that included A. phagocytophilum titre levels (results not shown) indicated no significant predictive effect of titre levels, but a significant effect (P < 0·001) of body weight when controlled for age.
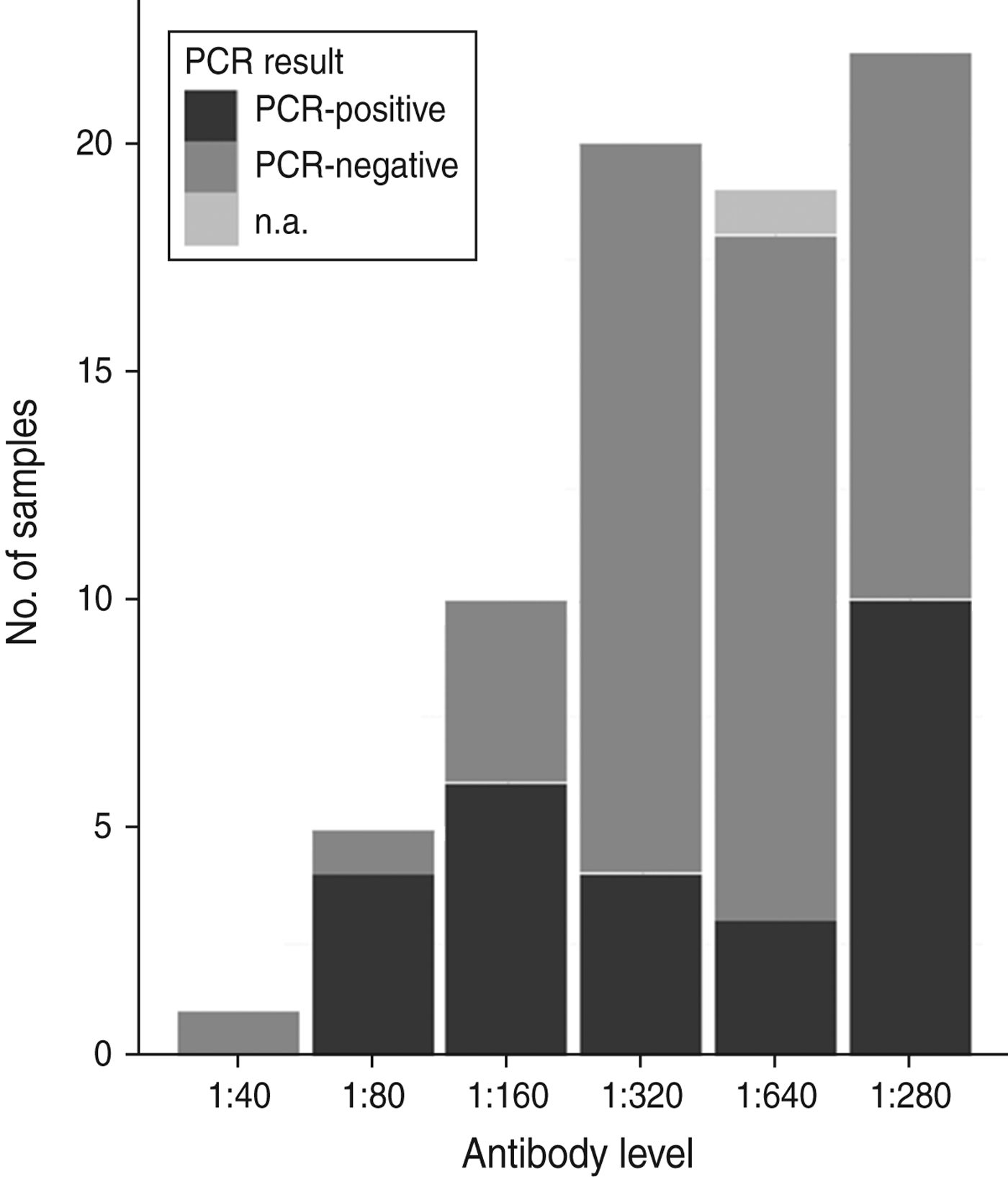
Fig. 2. An overview of numbers of samples with antibody titres and DNA presence of Anaplasma phagocytophilum in Swedish moose (Alces alces) sampled in 2009 and 2010. n.a., Not available.
The genetic sequencing revealed a 99% sequence similarity between the two samples for the 16S rRNA part. Furthermore, the island sample had 16S rRNA sequence similarities with an equine variant previously detected in Sweden (GenBank accession no. AY527214; 912/915 bp, 99·7%) [Reference Jäderlund27], a roe deer sequence from Austria (496/497 compared base pairs, 99·8%, GenBank accession nos. FJ812391–FJ81239 (type S) [Reference Silaghi13]). The groESL sequence from the island moose had a 99% sequence similarity (1267/1275 compared base pairs) with an HGA agent detected in humans in California (GenBank accession no. AF172163 [Reference Chae29]), unlike the mainland sample which shared 1252/1276 bp with the HGA agent (98·6%). Furthermore, the island groESL sequence was similar to a red deer and a roe deer sequence from Austria (530 bp compared, 99·6% and 99·4% sequence similarity, respectively, GenBank accession nos. GQ988767 and GQ988765, respectively) [Reference Silaghi13].
The 16S rRNA sequence from the mainland moose sample had a 99·9% sequence similarity with the equine variant reported previously from Sweden (GenBank accession no. AY527214; 914/915 bp [Reference Jäderlund27], and was identical (497/497 compared base pairs) to an Austrian roe deer sequence (GenBank accession nos. FJ812406–FJ812408, type Y) [Reference Silaghi13]. A shorter section (groEL) of the groESL gene sequence in the mainland moose sample showed a 100% sequence similarity (455/455 bp) with a roe deer A. phagocytophilum sequence detected in Austria (Gen Bank accession no. GQ988754), and a 99·8% sequence similarity (529/530 bp) with sequences found in both red deer and roe deer in the same paper [Reference Silaghi13] (GenBank accession no. GQ988763).
GenBank accession numbers for the novel moose sequences are KC800983 (island) and KC800984 (mainland) for the 16S rRNA sequences and KC800985 (island) and KC800986 (mainland) for the groESL sequences.
DISCUSSION
This is the first study on A. phagocytophilum infection in moose in Sweden, based on the presence of A. phagocytophilum DNA, and of antibodies. No similar large-scale study in cervids has previously been reported, apart from a Norwegian study of archived moose serum samples (collected 1994–2000), where a 43% seroprevalence in moose was found [Reference Stuen10]. In 2012 seroprevalence of A. phagocytophilum antibodies in 33 captured moose from a reported tick-infested area in southern Norway was 78% [Reference Milner and van Beest11]. Although the performance of the serological test in moose has not been validated, we consider the results to be reliable since the test used in three Norwegian studies [Reference Stuen10, Reference Milner and van Beest11, Reference Stuen30] is the same as the one used in this study. In addition, there are no reports of possible cross-reacting Anaplasma variants (e.g. A. marginale) in Sweden.
In the present study the overall prevalence of A. phagocytophilum DNA was 26·3%, with significant spatial difference between the island population (A) and the mainland populations (B, C, D). Spatial variation in prevalence of vector-borne pathogens has been described previously. Chaput et al. [Reference Chaput, Meek and Heimer31] reported spatial variation in incidence of the HE agent in humans in Connecticut, USA. In the present study, one reason might be the annual arrival of high numbers of tick-carrying migrating birds, which reportedly carry Ehrlichia sp. bacteria (HE agent [Reference Bjöersdorff32]). DNA in the present study also showed a temporal difference, with higher prevalence in 2008 and 2010, respectively. Temporal variation in A. phagocytophilum antibody prevalence has previously been reported by Rejmanek et al. [Reference Rejmanek33] in small mammals in California where the rate of exposure (antibody presence) varied over a 2·5-year period.
The range in A. phagocytophilum antibody titres (1:40–1:1280) observed, may be a result of several factors including the time of infection/exposure, re-infection, host immune response, variant of the bacterium, cyclic bacteraemia in the infected animal, and the sensitivity of the detection method [Reference Ladbury16, Reference Granquist34]. Antibodies against different Anaplasma variants have been shown to persist for at least 5 months in experimentally infected dogs (Ehrlichia sp. [Reference Egenvall35]), lambs (A. phagocytophilum [Reference Stuen, Olsson Engvall and Artursson36]) and horses (Ehrlichia phagocytophila [Reference Pusterla, Lutz and Braun37]), and for at least 2·5 months in red deer [Reference Stuen30].
The observed effect of A. phagocytophilum DNA presence on moose carcass weight described earlier in this report will not be further commented upon due to the small sample size (n = 77).
A 42·6% A. phagocytophilum DNA prevalence has been reported previously in 237 sampled roe deer in Denmark [Reference Skarphédinsson, Jensen and Kristiansen38]. Other European studies have shown varying A. phagocytophilum DNA prevalence ranging from 18% to 86% (roe deer [Reference Petrovec12–Reference Zeman and Pecha15, Reference Liz39, Reference Žele40]), 29–86% (red deer [Reference Petrovec12, Reference Silaghi13, Reference Žele40, Reference Polin41]), 50–72% (fallow deer, Dama dama [Reference Zeman and Pecha15, Reference Ebani42]), and 40% (sika deer, Cervus nippon [Reference Zeman and Pecha15]). These reports suggest that infection with A. phagocytophilum is common in cervids in Europe. However, more knowledge is needed about the pathogenesis, immune response, and clinical manifestation of A. phagocytophilum infection in order to assess the clinical and pathological significance of infection in free-ranging cervids such as moose. Experimentally infected Norwegian captive red deer (Cervus elaphus atlanticus) and reindeer (Rangifer tarandus) showed individual differences in clinical signs, time of seroconversion, and duration of antibody titre [Reference Stuen30, Reference Stuen43]. In reindeer, fever and neutropenia were noted, but not in red deer, other than a calf which showed a moderate increase in rectal temperature.
In the present study, antibodies against A. phagocytophilum were found in both A. phagocytophilum DNA-positive, and DNA-negative moose. The varying titres (1:40–1:1280) is an indication of when in time the animal is infected, or re-infected. Adding to this, no end-titration was performed which could have rendered even higher titres in some samples. Presumably, and as observed in sampled moose, the vectors (ticks) are probably active not only in the summer months but also during the autumn. Antibodies are reportedly known to persist longer than circulating bacteria, which could explain the observed discrepancy between seroprevalence and A. phagocytophilum DNA-based prevalence. This has been reported in a various A. phagocytophilum-infected ruminants including sheep [Reference Stuen, Olsson Engvall and Artursson36, Reference Paxton and Scott44] and horses [Reference Nyindo45]. Similar discrepancies have previously been reported in roe deer, where Zeman & Pecha [Reference Zeman and Pecha15] showed a 100% seroprevalence, and a 30% A. phagocytophilum DNA-based prevalence. In the same study, the discrepancy in mouflon was 100% and 3·5%, respectively. In wild boar from Slovenia, the discrepancy was 72% (serology) and 6·2% (PCR), respectively [Reference Žele40].
No association between antibody titre and A. phagocytophilum DNA presence was identified in this study (Fig. 2). Infection and host humoral immune response may be dependent on several factors, as stated previously. For instance, Stuen et al. [Reference Stuen46] reported that experimentally infected lambs had a detectable bacteraemia on several different occasions over a 13-week period. In the present study, both mainland and island moose had been exposed to A. phagocytophilum, but the A. phagocytophilum DNA prevalence varied between areas, and between sampling years (Table 1). A cyclic variation may also be present in moose infected with A. phagocytophilum, and if sampled on another occasion, such as during the summer months, the A. phagocytophilum DNA presence, and antibody prevalence, may have been different. The significance of body weight on the presence of A. phagocytophilum DNA was noted, and could be caused by a reduced weight gain, as reported by Stuen et al. [Reference Stuen, Bergström and Palmér47] in lambs infected on pasture. However, since this effect was not present in all sampled moose (sampled from 2007 to 2010), no conclusions have been drawn.
The results of the genetic sequencing A. phagocytophilum DNA-positive samples provide no evidence that moose are a host or a reservoir for different genetic variants. One of the sequenced samples from an island moose had genetic similarities with the HGA agent, which has previously been reported in Sweden [Reference Dumler5, Reference Bjöersdorff48]. More spleen samples from moose need to be sequenced before any further conclusions can be drawn.
In conclusion, this study demonstrated that moose from southern Sweden were commonly exposed to and infected with A. phagocytophilum. The A. phagocytophilum DNA prevalence varied between years (temporal variation) and between regions (spatial variation). Future studies should focus on the characterization of different genetic variants of A. phagocytophilum in free-ranging Swedish moose, and their pathogenic potential for moose and other mammalian species, such as domestic animals and humans.
ACKNOWLEDGEMENTS
The authors express their sincere gratitude to the hunters of southern Sweden for assistance in the sample collection. We also thank all colleagues at the Department of Pathology and Wildlife Diseases at the National Veterinary Institute (SVA) for their support in the handling of samples. We also thank the funding sources for their contributions to this study: the Swedish Environmental Protection Agency, the SVA, the Environmental Monitoring and Assessment programme, the Faculty of Veterinary Medicine and Animal Science at the Swedish University of Agricultural Sciences, and the Wildtech Project [European Union Seventh Framework Programme (FP7/2007-2013) grant agreement no. 222633].
DECLARATION OF INTEREST
None.






