INTRODUCTION
Aquatic environments have long been considered to be the reservoir of Vibrio cholerae, the causative agent of cholera worldwide [Reference Colwell and Huq1]. Identifying the source of a particular cholera epidemic and its transmission route is of significant importance for source-tracking and timely control of the cholera epidemic, thus preventing subsequent persistence of the epidemic [Reference Ghose2]. Moreover, better understanding of the microbiological behaviour of V. cholerae isolated from patients and water during dissemination, as well as the changes of its molecular virulence traits and genotypes, would be beneficial to health risk evaluation and management associated with active or accidental consumption of contaminated water [Reference Izadi3].
The survival and proliferation of V. cholerae under the influence of water temperature, salinity, pH, nutrients, biofilm formation, and amoebas has been investigated in the laboratory [Reference Singleton4–Reference Watnick6]. However, its survival behaviour in larger volumes of water, which better approximates the realistic conditions of natural waterways, is rarely studied. Since a lengthy cholera epidemic that occurred in 2005 in the Nansha District of Guangzhou indicated to us the probable existence of V. cholerae O1 Inaba in waterways, it was necessary to acquire information about the survival behaviour of representative epidemic strains in the large volume of waters sampled from epidemic sites.
Previous studies have paid attention to differences between virulence-associated genes of V. cholerae from various sources. Moreover, pulsed-field gel electrophoresis (PFGE), a commonly accepted epidemiological tool for investigating V. cholerae epidemics, has also provided information on the role of V. cholerae in both the environmental reservoir and cholera epidemic events [Reference Kam7, Reference Mahalingam8]. However, to the best of our knowledge, there are no reports examining the molecular variations of V. cholerae during a survival study utilizing a combination of virulence gene profile detection and PFGE fingerprinting determination.
Consequently, in the current study we first selected five representative strains from V. cholerae O1 and O139 isolated in 2005 after examining their virulence gene traits and molecular banding patterns. Next we observed the survival capacity of the five strains in large volumes of water sampled from waterways close to where the majority of cases were identified during the 2005 Nansha cholera outbreak. We then analysed the virulence-associated gene and genotypic changes of surviving isolates recovered from the survival test and as well as their role in cholera epidemics.
METHODS
Description of the study area and epidemic investigation
In 2005 a cholera outbreak lasting for nearly 3 months occurred in Nansha District, which lies at the estuary of Pearl River, the largest river in Guangdong Province, flowing from the northwest into the South China Sea. The waterway systems in Nansha District are the major source of irrigation, aquaculture and daily life for local residents, consisting of 21 parallel waterways in the Xinken area, six in Jibao Island and three in Longxue Island. After the emergence of cholera cases, an investigating team was sent to the local region for epidemic investigation and surveillance of the waterway system. We reviewed the findings from that investigation and then plotted geographical locations and an epidemic curve of cholera cases to describe the cholera epidemic.
Molecular identification of V. cholerae isolated in 2005 and strain selection for the survival test
This lengthy cholera outbreak prompted us to seek information about the survival behaviour of epidemic isolates in the large volumes of water sampled from epidemic sites. Accordingly, we prepared to choose representative O1 Inaba strains from the outbreak for the survival test, an assay for viable and culturable V. cholerae. For comparison, representative strains of O1 Ogawa and O139 were also incorporated. A total of 40 V. cholerae strains, consisting of 24 involved in the cholera epidemics and 16 from the environmental surveillance in 2005, were identified by polymerase chain reaction assay (PCR) and PFGE. Next, strains for the subsequent survival test were selected based on molecular epidemiological information obtained.
PCR
All the V. cholerae strains involved were subjected to monoplex PCR assay for virulence-associated genes. PCR primers specific for the virulence-associated genes ctxA, tcpA, ace, zot, encoding cholera toxin subunit A, toxin-coregulated pilus subunit A, accessory cholera enterotoxin, and zonula occludens toxin, respectively, were as described previously [Reference Singh9]. Amplification conditions were the same for the four gene fragments. PCR was performed with a PTC-220 thermal cycler (MJ Research Inc., USA) and in a 20 μl reaction mixture. Amplified products were separated in a 1% agarose gel.
PFGE
A modified protocol from the PulseNet rapid standardized one-day PFGE protocol for V. cholerae subtyping [Reference Cooper10] was employed for genotyping analysis. Briefly, plugs prepared from the mixture of V. cholerae suspensions and molten 1% SeaKem Gold gel (FMC, USA) were subjected to cell lysis and washing, followed by NotI enzyme digestion. After electrophoresis using the CHEF Mapper system (Bio-Rad, USA), the gel was then stained, and the image captured using the Gel Doc 2000 system (Bio-Rad). Profile processing, dendrogram construction and similarity analysis were conducted using the BioNumerics software package v. 4.0 (Applied Maths Inc., USA) with an optimization of 0·5% and a position tolerance of 1·2%. Any strain with more than one band difference was assigned a unique banding pattern (fingerprint or pulsotype).
Survival test
Large volumes of water used for the survival test were taken from epidemic sites of the 2005 cholera outbreak. Eight representative waterway sampling sites were selected. Seven of which were located in ‘Xinken area 15’ and ‘Jibao Island 4’ because the majority (57%) of the cholera cases were reported in these two sites. Another sampling site was chosen in Eastern Jibao Island because the waterway at this site was positive for O1 Inaba strain during the outbreak. At each site, numbered from 1 to 8, three replicate 20-litre water samples were collected from ~0·5 m below the water surface using 25-litre polypropylene containers. Within 3 h, all of the samples were sent to the laboratory, where three replicate 50 ml water samples were taken from three points distributed vertically, i.e. near the water surface, in the middle and near the bottom of each water container. The three replicate 50 ml water samples were mixed and examined for the presence or absence of V. cholerae O1 and O139.
A total of five selected strains of V. cholerae representing the most commonly seen V. cholerae serogroup or serotype in the Guangzhou area based on the molecular identification of 40 strains of V. cholerae were subjected to this survival assay. A suspension of the selected strains was made and adjusted to 1·0 McFarland turbidity (about 1·0 × 108 c.f.u./ml), simulating the amount of V. cholerae shed by a cholera patient. One millilitre of each bacterial suspension was pipetted into unsterile water in a 25-litre container. In the case of water sample nos. 1, 2 and 8, suspensions of the same strain were added. For samples from the other five sites, a combination of three different V. cholerae strains was added into each of three replicate 25-litre containers from a single site. The following day after strain inoculation, three replicate 50 ml water samples were collected from each of the 18 numbered containers and mixed as described above. Isolation and identification of V. cholerae O1 and O139 was performed for each mixed sample. The same procedure was repeated 3 days after inoculation and then every 7 days from that day on. The survival test continued until the water sample with the longest detection period was negative for culturability for three consecutive samplings. In addition, for each sample positive for culturability, five randomly selected surviving isolates from the culture plate were picked up and stocked for subsequent virulence gene and fingerprint analysis by PCR and PFGE, respectively.
Isolation and identification of V. cholerae O1 and O139
To isolate and identify V. cholerae serogroups O1 and O139 from water taken in the survival test, each of the water samples was subject to enrichment; first, by adding 10 ml of 10 × alkaline peptone water (APW, pH 8·6) to a 90 ml water sample. After incubation at 37°C for 6–8 h, the enriched culture was streaked on thiosulfate citrate bile sugar (TCBS) agar and incubated at 37°C for 8–12 h. Presumptive yellow colonies were randomly selected, pure cultured on nutrient agar plates and then detected by the slide agglutination test with V. cholerae O1 and O139 polyvalent and monovalent V. cholerae diagnostic antisera (Denka Seiken Co. Ltd, Japan).
RESULTS
Description of the 2005 cholera outbreak
According to the data available from the epidemic investigation in Nansha District, the cholera outbreak lasted for up to 82 days from 19 September to 9 December 2005. Thirty-seven diarrhoea patients with symptoms varying in severity from mild to severe cholera were reported with an attack rate of 0·046%. The epidemic curve depicted all the cholera cases by date of onset and indicated a continuing exposure of nearly 3 months (Fig. 1). Of note, the epidemic curve also showed clustering of cases during different time periods from different sites. In the first phase, 11 cholera cases were reported from 19 September to 2 October and distributed at ‘Xinken 11–16’. In the second phase, 14 cases were distributed from 18 to 30 October at ‘Jibao Island 4 and 5’. In the third phase, 12 cases re-emerged from 6 November to 9 December at ‘Xinken 11–16’ as well as at Longxue, Masha, and Pearl Farm areas. Geographical distribution of the cholera outbreak (Fig. 2) indicated that 16 cases were distributed at ‘Xinken waterways 11, 14, 15 and 16’, while 17 cases were distributed at ‘Jibao Island waterways 3–5’. ‘Xinken 15’ and ‘Jibao Island 4’ had the largest number of cases. The cases included 17 males and 20 females and 11/37 cases belonged to four families. From the majority of these patients, V. cholerae O1 Inaba was isolated and considered the causative pathogen of this outbreak. Moreover, Inaba strains were isolated from the waterways of ‘Jibao Island 4’ and Eastern Jibao Island (Fig. 2). Epidemic investigation also showed that all cholera cases lived near the waterways and/or had recent farm-work histories nearby. With the exception of one case living in a fishing boat and another in a working vessel, nearly all of the cholera cases lived near the bank of the waterways. Most of these individuals took water directly from the waterways for kitchen use, domestic and personal hygiene. In addition, basic toilets constructed near waterways often released ineffectively treated faecal matter into the waterways. All these epidemic investigations, combined with laboratory confirmation of V. cholerae O1 Inaba strains from the cholera cases, added to the possibility that this was a waterborne cholera outbreak that was elicited by and transmitted through the waterways around this region.
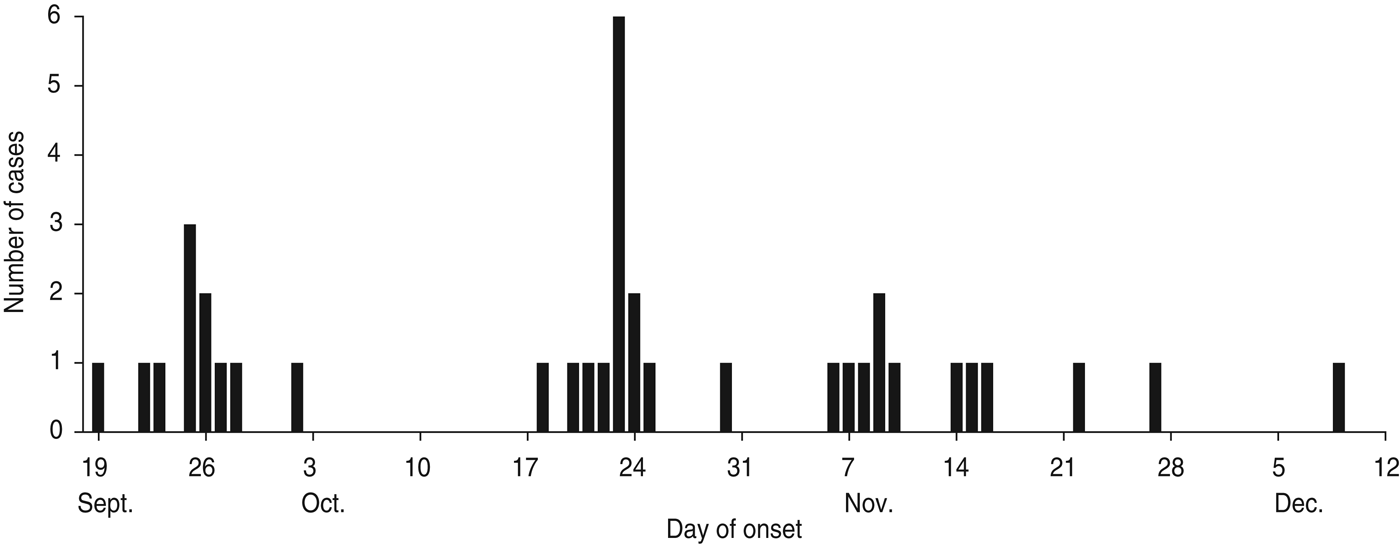
Fig. 1. Epidemic curve of reported cholera cases by date of onset (n = 37), Nansha District, Guangzhou, China, 19 September to 10 December 2005. Temporal distribution of cholera cases were as follows: phase 1 (19 September to 2 October); phase 2 (18 October to 30 October); phase 3 (6 November to 10 December).

Fig. 2. [colour online]. Map showing the geographical distribution of strains of Vibrio cholerae used in the study. (a) Map of China. (b) Map of Guangzhou city in Guangdong Province. (c) Geographical location of the cholera outbreak in Nansha District. Cholera cases distributed in each epidemic site are represented by histograms with the number of cases at the top. The waterways ‘Jibao Island 4’ and Eastern Jibao Island, where O1 Inaba strains were isolated, are each symbolized by a flag.
Virulence-associated genes and pulsotypes of V. cholerae strains isolated in 2005
Virulence-associated genes were determined by PCR for all 40 V. cholerae isolated in 2005 from cholera epidemic cases and environmental surveillance. Nearly all of the 24 epidemic-associated strains were found to be positive for the genes ctxA, tcpA, ace and zot, except for one O1 Inaba strain 05XB0939 isolated from Eastern Jibao Island during the cholera outbreak. By contrast, another O1 Inaba strain 05XB0940 isolated from the adjacent ‘Jibao Island 4’ waterway carried the four virulence-associated genes (Table 1). Of 16 strains from environmental surveillance, one strain was positive for all four genes, while others were negative for either both ctxA and tcpA or all the four genes (data not shown).
Table 1. Epidemiological data and virulence-associated gene profiles of Vibrio cholerae strains isolated in 2005
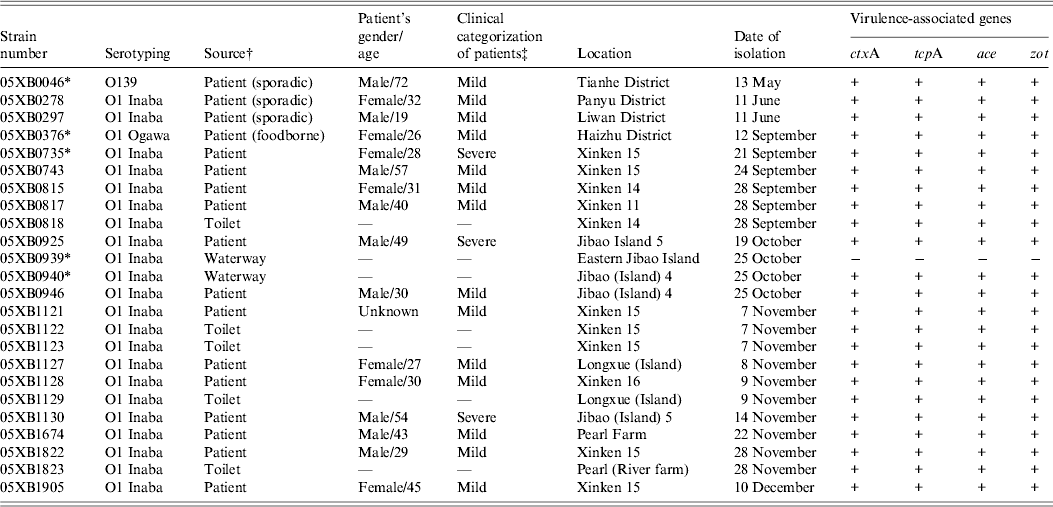
* Indicates strains selected for the survival test.
† The strains involved in the epidemic were isolated from the 2005 cholera outbreak unless indicated in parentheses.
‡ ‘Mild’ indicates patients suffered from painless diarrhoea, usually without vomiting; ‘severe’ indicates patients suffered from dehydration and electrolyte imbalances.
PFGE analysis indicated that 13 strains of V. cholerae O1 Inaba from cases during the cholera outbreak exhibited three slightly distinct pulsotypes A, B and C (>96·8% similarity), differing by one or two bands (Fig. 3). Interestingly, strain 05XB0940 from ‘Jibao Island 4’ waterway during this cholera outbreak shared the same pulsotype A with some of the strains from the outbreak patients. By contrast, the waterway strain 05XB0939 from the adjacent Eastern Jibao Island was epidemiologically unrelated to strains from outbreak patients (76·8% similarity). In addition, all of the 16 strains from environmental surveillance were obviously heterogeneous genotypically, with 12 different pulsotypes identified (data not shown).

Fig. 3. PFGE dendrogram of Vibrio cholerae strains involved in the Nansha cholera outbreak in 2005.
Strain selection for the survival test
Based on the molecular identification of 40 strains of V. cholerae, a panel of five strains representing the most commonly seen V. cholerae serogroup or serotype in the Guangzhou area were selected for the survival test. They included three O1 Inaba strains (05XB0735, 05XB0939, 05XB0940), one O1 Ogawa strain (05XB0376) and one O139 strain (05XB0046) (Table 1). The O1 Inaba strain 05XB0735 from the first reported case was selected as a representative strain of the Nansha outbreak and the likely dominant pathogenic clone (pulsotype B) epidemic in 2005. One selected O1 Inaba strain 05XB0940 from ‘Jibao Island 4’ was very likely to be involved in the outbreak because it carried the four virulence-associated genes and showed the same pulsotype to some of the patient isolates (Fig. 3). Another Inaba strain 05XB0939 isolated from neighbouring Eastern Jibao Island and negative for the four genes was also selected for comparison.
Strain survival (culturability)
After addition of the five strains into 18 tanks of large-volume water samples (Table 2), culturable V. cholerae was detected from all of the water samples 1 and 3 days later. However, after 10 days, 11 of the 18 water samples were positive for culturability, and 17 days later the number positive for culturability increased to 15. Half of the inoculated samples were still positive 24 days later. Thirty-one and 38 days after addition of the strains, the remaining number of water samples with survival positivity fell to four and two, respectively, with no strains recovered for three consecutive weeks after 45 days. Strains 05XB0735 and 05XB0940 in some of the water samples exhibited strong survival capacity up to 38 days. In addition, for half of the water samples, V. cholerae was detected initially, followed by a negative result for culturability, it was then detected again, although it was ultimately non-culturable.
Table 2. Survival of Vibrio cholerae in water samples after designated intervals
+, Survived; –, not detected.
When surviving isolate were recovered from a water sample, they were assigned a new number. For example, when experimental strain 05XB00735 was added to water sample 2-1 and was detected 10 days later, the surviving isolate was designated as 2–735(3).
Molecular identification of the surviving isolates of V. cholerae recovered from the survival test
All surviving isolates were identified by PCR and PFGE to observe their molecular changes after the survival test. PCR showed that virulence-associated gene profiles of the surviving isolates were the same as their original strains and appeared to undergo no changes. A panel of 15 representative surviving isolates was used to construct a dendrogram with the five original strains following PFGE. They were grouped into three main clusters (Fig. 4). One surviving isolate, 6–940(7), derived from 05XB0940 shared the same fingerprints (pulsotype B) with strain 05XB0735, the first outbreak patient strain. Unexpectedly, the banding pattern of one surviving isolate, 8–735(4), from O1 Inaba 05XB0735 was indistinguishable to that of O1 Ogawa strain 05XB0376. No genotypic changes were observed in the fingerprints of 05XB0376 and its surviving isolates. Further, the O1 Inaba strain 05XB0939 and O139 strain 05XB0046 underwent minor variations from their original strains (Fig. 4).

Fig. 4. PFGE dendrogram of five strains of Vibrio cholerae and the representative surviving isolates of V. cholerae recovered from the survival test. They were grouped into three main clusters, indicated by black circles, stars and squares, respectively.
DISCUSSION
In this study, through epidemic investigation and laboratory testing of the likely waterborne cholera outbreak in Nansha District in 2005, we had the following main findings: (1) outbreak-related V. cholerae strains survived for a substantial period of time in water sources sampled near the outbreak sites, although not as long as the entire length of the outbreak period; (2) important pathogenic genes were preserved during survival in these waters; (3) variations in PFGE fingerprints occurred over time; (4) PFGE patterns of certain V. cholerae strains of different serotypes changed to become more alike while cultured in outbreak waters.
The majority of the strains in the survival test exhibited an average survival period of 2–4 weeks. Some of them were able to survive in sampled waters for up to 38 days. Their relatively strong survival capacity seems to parallel the clustering of cases depicted by the epidemic curve. Comparison with a previous study that V. cholerae O1 in microcosms became non-culturable on TCBS agar within 10–15 days [Reference Alam11] seems to indicate that culturability of V. cholerae in a large volume of water is a little higher than that in laboratory microcosms. On the other hand, the strains failed to persist for an extended period of time, probably because unfavourable temperature and/or nutrient deprivation make V. cholerae resort to an alternative survival strategy, a ‘viable but non-culturable (VBNC) state’ [Reference Chaiyanan12]. The intermittent positivity in culture observed in half of the water samples in the current study might be suggestive of the beginning of the VBNC state. Moreover, unculturability may also be related to the fact that V. cholerae freshly discharged into the environment is significantly less robust than cells adapted to environmental conditions. At the in situ cholera epidemic field sites, in vivo-formed biofilm-like clumps of V. cholerae shed by cholera patients may extend the culturability of V. cholerae in water and probably contribute to enhanced infectivity and environmental persistence of pathogenic V. cholerae [Reference Faruque13]. Consequently, V. cholerae shed by patients probably persisted in the water due to its considerable surviving potential, and the re-emergence of cases was contingent upon the availability of an appropriate transmission route, such as contact with epidemic water.
The genes ctxA and tcpA, comprising the virulence gene cassette which is necessary for intestinal adherence and colonization, were critical for pathogenicity of V. cholerae. All of the toxigenic strains examined in the survival test experienced no changes in virulence-associated genes, maintaining pathogenic potential in the aquatic environment, which might be one of the reasons for the extended cholera epidemic in Nansha District. Even though some researchers suggest that the expression of a factor that enhances environmental persistence actually decreases virulence [Reference Watnick6], and that environmental conditions could affect expression of virulence genes in V. cholerae [Reference Castaneda14], such phenomena were not observed in the virulence gene profiles of the culturable surviving isolates in the present study, probably because genetic changes occur over a longer period of time, in a manner similar to evolution [Reference Chakraborty15, Reference Faruque, Albert and Mekalanos16].
To probe the minute variation of PFGE fingerprints of V. cholerae in this study, a strain with one band difference was designated as an independent pulsotype. Although no previous study has utilized PFGE to study the changes of molecular subtypes of V. cholerae during a survival test, we found that slight variations of PFGE fingerprints occurred in four of the five original strains in the survival test. This seems to suggest that chromosomal genomes of V. cholerae are prone to change in an aquatic environment. Such changes might be due to loss or gain of a restriction site on the genome. However, whether this confers survival advantage needs further study. We also had an unexpected finding that a recovered surviving isolate of 05XB0940 from the survival test mutated to share the same PFGE pattern to that of the first patient. Such a result indicates that a pathogenic clone of V. cholerae shed by patients into the waterways might be able to mutate into similar clones through limited variation in molecular fingerprints and maintain virulent potential. Since the majority of local residents lived near the waterways and took water directly from the waterways for kitchen use, domestic and personal hygiene, their continuous exposure to contaminated epidemic waters were evident. Coexistence of slightly different pathogenic clones of V. cholerae may have then caused an intermittent re-emergence of cases.
Unexpectedly, this study found that strains of distinct serotypes with slightly different PFGE fingerprints (Inaba and Ogawa strains) could evolve to share identical PFGE fingerprints, with serotypes and virulence gene profiles remaining unchanged. This is a unique observation in a survival test. Derivation into indistinguishable pulsotypes without serotype conversion in the survival test might be due to the absence of changes in the wbtT gene, a gene determining serotype switching in the water environment, instead of immune selection in infected cholera cases [Reference Ito17, Reference Stroeher18].
The laboratory-based simulation performed in the present study does not necessarily replicate actual epidemic events during several months in the epidemic field site. Thus, our findings cannot exclude the possibility that V. cholerae may undergo other genetic or genotypic changes. Moreover, it is difficult to define the influence of fingerprint changes on survival capacity of V. cholerae under study. V. cholerae positive for ctxA and tcpA may persist in waters long enough and undergo limited variations in genomic DNA, thus maintaining its potential to cause disease again. The survival capacity of V. cholerae in an aquatic environment in the epidemic field is likely to be higher than that observed in this study, which might account for the intermittent re-emergence of cholera cases in the Nansha outbreak.
ACKNOWLEDGEMENTS
This research was supported by the Guangzhou Medical and Hygienic Research Fund (grant nos. 2005-YB-113, 2009-YB-121) and the Medical Research Fund of Guangdong Province (grant no. A2010493).
DECLARATION OF INTEREST
None.









