INTRODUCTION
Campylobacter enteritis is one of the most frequent causes of diarrhoea in the United Kingdom with 39 745 cases reported in 2004 [1]. Despite this burden of human infection, the epidemiology is yet to be entirely understood with the transmission route to humans and into the food chain still not fully ascertained [Reference Gillespie2, Reference Gillespie3]. Human infection results in gastroenteritis with symptoms ranging from mild to severe inflammatory diarrhoea, dependent on the infecting strain and the host response [Reference Ketley4, Reference Butzler5]. Laboratory diagnosis of Campylobacter gastroenteritis is by cultural isolation of Campylobacter from faeces, which can take 2–3 days and will only identify the causative organism as Campylobacter spp. [Reference Maher6–Reference Lastovica and le Roux8]. Many typing methods have been described for Campylobacter [Reference Ribot9–Reference Dingle18], however, very few can be carried out quickly and enable strain identification directly from a faecal sample without a prior culturing step. More rapid and effective methods for the detection of specific types of campylobacters from faecal samples may facilitate a better understanding of the epidemiology and help to further clarify the sources of human infection.
The high-resolution genotyping technique, multilocus sequence typing (MLST) [Reference Dingle18] has contributed to our understanding of the population biology of the most commonly isolated species C. jejuni, by identification of lineages or clonal complexes, containing groups of closely related strains [Reference Dingle19]. The C. jejuni clonal complexes have been recognized as potential epidemiological groupings with possible host associations [Reference Dingle19, Reference Colles20]. To facilitate rapid clonal complex recognition, a strategy for identification of six major clonal complexes associated with human infection has been developed, based on the presence of specific single nucleotide polymorphisms (SNPs) within the MLST scheme alleles [Reference Best21]. The strategy uses a real-time PCR platform (Taqman, Applied Biosystems, Warrington, UK) and utilizes allelic discrimination assays to accurately determine the presence of SNPs, which in specific combinations are diagnostic for six clonal complexes. Our objective here was to use the MLST SNP-based assays for the direct detection of C. jejuni by clonal complex from human faecal specimens, and then confirm the accuracy of the clonal complex designation from the SNP-based assays by performing MLST on the cultured faecal material.
METHODS
Clinical specimens
Faecal specimens obtained from patients with symptoms of gastroenteritis (n=101) were collected over a 6-month period in 2004 from the Microbiology Laboratory at the Royal Preston Hospital and Clinical Microbiology laboratories in Manchester. These were either microbiologically proven cases of Campylobacter infection (n=70) or of other gastrointestinal infection (n=31) of known aetiology including Giardia spp. (12), Cryptosporidium spp. (7), Salmonella spp. (11) and Vibrio parahaemolyticus (1). Upon receipt at the Manchester Health Protection Agency laboratory, the samples were transferred to a Category III Enteric Microbiology Laboratory and a 1 ml aliquot transferred to sterile 2 ml tubes and stored at −20°C.
Direct DNA isolation from faeces
Chromosomal DNA was extracted from a total of 103 faecal specimens including two negative control samples obtained from healthy people with no symptoms of gastroenteritis using the Qiagen Qiamp DNA Mini Stool kit (Crawley, UK). Extracted DNA samples were stored at −20°C until required.
Bacterial reference strains
DNA extracts from C. jejuni (NCTC 11168) and C. coli (NCTC 12110) were used as species controls. DNA extracts from MLST reference strains [Reference Wareing22] corresponding to the six clonal complexes were used as controls for the SNP assays.
C. jejuni/C. coli identification from faecal samples
DNA extracts (diluted 1/10) were tested for C. jejuni or C. coli using a previously described Taqman assay [Reference Best23] with primers and probes for the genes ceuE (for C. coli) and mapA (for C. jejuni).
SNP assays for MLST clonal complexes on faecal samples
All DNA extracts from faecal specimens (n=103) were tested with the SNP assays identified as specific for one of the six major clonal complexes (Table 1). Real-time PCR assays were performed using the Applied Biosystems SDS 7000 in a total volume of 25 μl including 2·5 μl (diluted 1/10) DNA extract, 300 nm forward and reverse primers, 100–200 nm minor groove binding (MGB) probes (Applied Biosystems, Warrington, UK) and 1× Taqman universal mastermix. Cycling comprised 10 min at 50°C, 10 min at 95°C, followed by 40 cycles of denaturation at 95°C for 1 min and annealing/extension at 60°C for 1 min. SNPs were detected directly by monitoring the increase in fluorescence and clonal complexes indicated by the presence of SNPs in specific combinations. Results were displayed as a C T (threshold cycle) where the presence of the informative SNP was recognizable with a C T number within the range 14–25 and a signal strength ΔR n>1.
Table 1. Single nucleotide polymorphisms (SNPs) used for identification of each clonal complex

MLST sequencing from bacterial cultures
The original isolates from the faecal specimens were obtained for typing by MLST, in order to establish whether the clonal complex identified by the SNP strategy was correct. The cultured isolates (n=51) included all the samples testing positive for a clonal complex by the SNP assays in addition to a random selection of samples which were negative for any SNPs. Chromosomal DNA was prepared with the Roche MagNApure (Roche Diagnostics, Lewes, UK) using the Total Nucleic Acid Extraction kit, according to the manufacturer's instructions.
MLST was performed according to the method described by Dingle et al. [Reference Dingle18, Reference Dingle24] using published primers for C. jejuni or C. coli where required. DNA sequencing was carried out in forward and reverse directions and products separated on a Beckman CEQ 8000 capillary sequencer (Beckman, High Wycombe, UK). Contigs were assembled, trimmed and aligned using BioEdit (Tom Hall, Ibis Therapeutics, Carlsbad, CA, USA). All alleles, sequence types (ST) and clonal complexes were assigned by use of the Campylobacter MLST website (http://pubmlst.org/campylobacter).
RESULTS
Direct DNA isolation from faecal specimens
DNA extractions were performed successfully using the Qiagen QIAamp DNA Mini Stool kit from 103 faecal specimens. Sensitivity of the SNP assays was tested by using spiked faecal samples obtained from healthy people with no signs of gastroenteritis. An average sensitivity of the panel of SNP assays direct from the faecal specimens using this extraction method was calculated as 50 c.f.u. per PCR reaction.
Direct species identification from faecal specimens
The Taqman assay for identification of species was carried out on the neat DNA extracts (101) obtained directly from faeces. This identified 45 samples positive for Campylobacter of which 43 were C. jejuni and two were C. coli. In order to improve the detection rate, a range of dilutions of the DNA was carried out in order to prevent any inhibition within the PCR reaction. Dilutions (10−3) of the faecal specimens enabled further samples (70) to be identified as positive for Campylobacter of which 68 were C. jejuni and two were identified as C. coli. C T values were within the range of 27–35 and with a signal strength of ΔR n>1·0. In addition, four samples, previously unidentified as Campylobacter spp., were also shown to be positive for C. jejuni by the assay for species. These samples were microbiologically identified as Giardia spp. (1), Cryptosporidium spp. (2) and Salmonella spp. (1) (C T values of 30, 32, 30 and 34 respectively and ΔR n>1·0). Also one sample, which had previously been identified as Salmonella spp., was identified as C. coli (C T value 30, ΔR n>1). The remaining 26 samples were negative for C. jejuni and C. coli (C T values>35 and ΔR n<1).
Direct detection of specific C. jejuni SNPs by real-time PCR
All samples (101) were tested with the whole panel of SNP assays in order to detect specific combinations of SNPs, which were indicative of the six major clonal complexes (Table 1) [Reference Best23], however only the C. jejuni (68) confirmed samples were expected to provide a result. C T values obtained for the Taqman assays when a SNP was present were within the range of 20–33 and with a signal strength of ΔR n>1·0. Where no SNP was present a C T value of 40 and signal strength ΔR n<0·2 was seen. For example, isolate 38 (Table 2) was identified as belonging to clonal complex ST-21 with the presence of the three SNPs glnA1 108 (C T=26, ΔR n=1·1), glnA1 267 (C T=28, ΔR n=1·3) and tkt_1 330 (C T=26, ΔR n=1·2).
Table 2. MLST clonal complex results and SNP clonal complex results for C. jejuni isolates
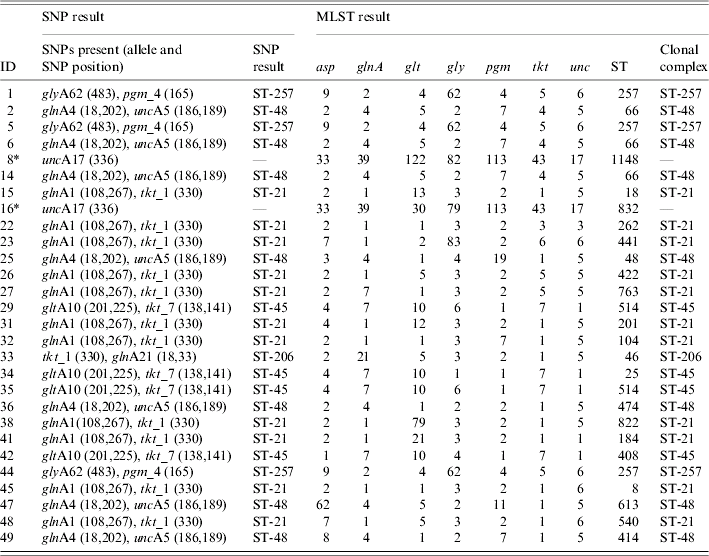
MLST, Multi locus sequence typing; SNP, single nucleotide polymorphisms; ST, sequence typing.
* C. coli isolates tested which were positive for the unc17 SNP, but not assigned to a clonal complex.
In total, direct testing from the faecal specimen identified 26 (38%) of the C. jejuni-confirmed samples as belonging to one of the six major clonal complexes (Table 3). Of these, 11 (42%) samples were assigned to clonal complex ST-21, seven (27%) samples to clonal complex ST-48, three (12%) samples to ST-257, one (4%) sample to ST-206 and four (15%) samples to ST-45.
Table 3. Distribution of C. jejuni samples positive for one of the six clonal complexes by each method

MLST, Multilocus sequence typing; SNP, single nucleotide polymorphisms.
* Tested on faecal extract.
† Tested on isolate.
The two C. coli isolates possessed the SNP unc17 333 (indicative of clonal complex ST-61 in C. jejuni). In three samples (5, 14, 25) (Table 2) the presence of SNPs specific for two clonal complexes were present. In these cases the clonal complex was assigned based on the SNPs, which were present in greater quantity (by lower C T value). This was possible due to the quantitative capability of the Taqman instrument. A lower C T value was indicative of a greater quantity of target DNA present. For example in sample 25, SNPs indicative of two clonal complexes (ST-48 and ST-257) were present, C T values for the SNPs indicative of clonal complex ST-48 were lower, e.g. 29 (glnA4 18), 29 (glnA4 202) and 28 (unc_17), when compared to the values for ST-257, e.g. 33 (glyA62 438) and 33 (pgm_4 165). Some of the culture-confirmed Campylobacter-negative samples also showed the presence of SNPs, but not in specific combinations to be able to recognize a clonal complex.
The four samples, which had been identified as C. jejuni, but microbiologically, confirmed as other species were positive for some SNPs. The sample which was microbiologically confirmed as Giardia spp. but positive for C. jejuni by the speciation assay partly possessed the SNPs for clonal complexes ST-21 [glnA1 267 (C T=31) and tkt_1 330 (C T=30)], of the two Cryptosporidium spp. isolates one partly possessed the SNPs for clonal complexes ST-206 and ST-257 [glnA21 18,33 (C T=26) and pgm_4 165 (C T=27)] the other partly possessed SNPs for ST-21 [glnA1 108 (C T=28) and tkt_1 330 (C T=26)] and the Salmonella spp. isolate partly possessed the SNPs for ST-206 [glnA 21 18,33 (C T=32)]. This was suggestive of C. jejuni presence, however, the lack of SNPs in specific combinations to be able to recognize a clonal complex and also the lack of culture would not be sufficient evidence to confirm the presence of C. jejuni in these extracts. The sample, which had previously been identified as Salmonella spp., but showed a positive reaction for C. coli with the Taqman assay for species, was negative for all the SNP reactions.
Confirmation of clonal complex by MLST from the cultured specimen
Twenty-six (38%) samples, which had been identified as belonging to one of the six clonal complexes by the SNP strategy, were confirmed correctly using MLST from the cultured specimen. Table 3 shows the MLST results for the 26 C. jejuni isolates identified by the SNP strategy as belonging to a clonal complex. A proportion (23) of SNP clonal complex negative samples were also tested by MLST to confirm that none of the six major clonal complexes had been missed by the SNP approach. These isolates tested were identifiable as other clonal complexes e.g. ST-573, ST-353, ST-42, ST-22 and ST-443, which would be unidentifiable by the SNP strategy, or were unassigned to a clonal complex. None were identified as belonging to one of the six clonal complexes identifiable with the SNP strategy. The C. coli isolates had been identified as having the SNP uncA17 indicative of ST-61 in C. jejuni, this was confirmed by the presence of the allele uncA17 by using C. coli-specific published primers [Reference Dingle24]. For the isolates where the SNPs were present in combinations indicative of more than one clonal complex, MLST confirmed the clonal complex identified in the highest proportion.
DISCUSSION
Our approach showed that it was possible to detect specific C. jejuni strains assigned to one of the six major clonal complexes, directly from human faecal specimens from infected patients. This strategy represents a new rapid approach for real-time PCR to be used for detection and rapid characterization. All existing real-time PCR methods are used for detection of species-specific genes used for the identification of bacterial pathogens, however, this strategy is capable of going one step further, and enables the detection of particular strains of Campylobacter based on established genetic lineages directly from faecal specimens. The extraction system used is a specifically designed kit for the production of DNA from samples, which is free from PCR inhibitors, of good quality and suitable for sequence detection. Previous methods for direct DNA extraction have been lengthy procedures often utilizing toxic chemicals and also often requiring a second purification step to enable successful downstream processing by PCR [Reference Boom25, Reference McOrist, Jackson and Bird26]. The rapidity of this approach enables a result to be obtained quickly if an urgent diagnosis is required, or rapid confirmation in the case of a suspected outbreak. It also could be used to link outbreak cases together or potentially link human cases with samples of contaminated food products.
The Taqman assay for species was originally designed and has previously been tested for the identification of C. jejuni or C. coli from cultured isolates. Testing directly on extracts from faecal specimens has demonstrated that it may be insensitive, as a range of dilutions had to be performed to remove any potential inhibitors and the resulting C T values were higher than expected. The SNP assays appeared to be more specific for C. jejuni, as C T values were lower, suggesting a good quantity of DNA was present. The improved sensitivity of the SNP detection may be due to the MGB probes used in the reaction, which offer an increased level of sensitivity over the standard Taqman probes used in the assay for species. Also this assay identified either C. jejuni or C. coli in a number of samples, which had previously been identified as other organisms. Initially this was assumed to be contamination, however, the presence of SNPs within these samples suggests that there may be a genuine presence of Campylobacter mixed with other organisms but further evidence would be required to substantiate this finding. Mixed organism infections of C. jejuni and Giardia spp. [Reference Chunge27] or Cryptosporidium spp. [Reference Duke28] and C. coli and Salmonella spp. [Reference Enzensberger29] have been documented before. This is being investigated and further work needs to be carried out to verify whether the assay is sufficiently sensitive and also with respect to any possible sequence variation, which may be present in these strains tested. Interestingly, it was possible to imply the species designation of the samples with the SNP assays. Although not designed for identification of species, the presence of the SNP (uncA17) implied the C. coli species tested in the study and the presence of the remainder implied C. jejuni species. This provided a useful confirmatory test, however, this could not be relied upon alone as a test for species and further work is required on a larger scale to verify this.
There are a few limitations to the SNP approach described. First, the full complement of MLST strain typing data such as the allelic profile and sequence type are not obtained by the SNP approach, and only the clonal complex is identified. For the purposes of quickly screening isolates this typing information may be adequate, however, for more detailed studies of population genetics or epidemiological investigations, further resolution than the clonal complex may be required. Additionally, the current range of clonal complexes identified may not be appropriate for testing other sample populations, for example water or environmental samples where different clonal complexes may prevail in greater proportions. New sequence types may go undetected as a result of using the SNP approach. However, the SNPs used are highly specific for the six described clonal complexes as tested on human samples, therefore any new types would not be identified by the SNPs and would require MLST. The proportion of isolates identified (38%) in this sample set was sufficient in order to make a substantial difference to the numbers if MLST were to be the next stage in identification.
The nature of this type of strategy raises new problems, not encountered before due to the limitations of the existing techniques. Of interest is the ability to detect potential mixes of clonal complexes. Three samples were identified as having the SNPs present for more than one clonal complex, which in this reaction format is possible. Whether this is genuine, is very difficult to establish. MLST by PCR and sequencing does not provide a reliable indicator of mixes and also does not provide the required level of sensitivity. Capillary sequencing is only able to read one base at any position even if a mix is present. Any double base may be a result of a poor sequence read, as the Beckman sequencing chemistry has a tendency to insert bases into a read if the signal during the sequencing reaction is particularly low and the presence of genuine mixes (by the presence of two bases) would be difficult to detect. To establish that the SNP strategy was capable of detecting mixtures, artificial mixed suspensions were created by mixing equal and varying quantities of different clonal complexes together and then tested using the SNP assays. This showed that it was possible and also demonstrated the quantitative ability of the Taqman assays for detecting the SNPs present in greater quantity (by lower C T value). Also during this investigation single colonies were picked from the culture plates to ensure that mixtures were not being taken. Further work is required to establish that either genuine mixes do exist, or that the mixed results are due to contamination.
The MLST SNP approach provides potential advantages over existing typing schemes and has many of the merits of MLST. The data is portable allowing easy transfer between laboratories and is also amenable to electronic storage. Real-time PCR equipment such as the Applied Biosystems SDS 7000 and associated chemistries are increasingly more readily available, and the primers and probes used in this strategy could be transferable to other real-time PCR platforms with equal success. Additionally, the results can be compared with MLST results already obtained and listed in the database, this provides for the SNP MLST approach to be used as a complementary system to MLST, enabling the rapid and effective identification of common clonal complexes. The greater sensitivity of the Taqman system makes it possible to detect Campylobacter DNA present, which could be missed by conventional PCR. Also it is possible that if very low numbers of Campylobacter cells were present then growth in culture may not be possible. The direct SNP approach enables potential detection of Campylobacter, which may be missed during culture, due to the sublethally injured organism being exposed to harsh culture conditions within selective media. The SNP approach gives a different perspective on the presence of Campylobacter directly from faecal specimens.
There are many associated and recognized SNP papers [Reference Gutacker30, Reference Wahab31] but no previous studies to date have investigated the concept of using a SNP-based typing approach for the identification of specific Campylobacter strains. Typing by SNPs based on the data generated from MLST databases has been described for other organisms including N. meningitidis and S. aureus [Reference Robertson32, Reference Stephens33] however these approaches have been complex systems used to identify the sequence type of an isolate by multiple PCRs. No method has been described for the use of SNPs for identification of Campylobacter MLST clonal complexes, and it is the first description of a real-time PCR approach for the direct detection and further strain identification for this organism. Additionally, the ability to use the method directly on faecal specimens enables a rapid approach to real-time epidemiological studies. At present there is no method for the accurate identification of Campylobacter isolates to strain level directly from faecal specimens. This would aid epidemiological studies and could be used to give an indication of the specific types of campylobacters present within clinical samples.
Successful application of the strategy for the six major clonal complexes associated with human infection as described here suggests that the scheme could be extended to encompass other clonal complexes of the C. jejuni MLST scheme and to accommodate changes in prevalence of particular strains. Additionally, with the expansion of the MLST scheme to incorporate other Campylobacter species such as the second most commonly isolated species C. coli [Reference Dingle24] it would be possible to extend the SNP scheme to enable identification of other campylobacters. It has been demonstrated that one of the SNPs which has been used to identify the ST-61 clonal complex can also be used to identify the C. coli isolates within this dataset, suggesting that it would be possible to use the same SNPs for other species. The strategy could be effective for the rapid identification of specific strains, however, it has not been developed as a replacement for MLST. To provide the strain coverage achieved by MLST using a SNP approach would involve multiple reactions encompassing large numbers of primers and probes and complex reactions to cover all the clonal complexes. If the whole MLST clonal complex coverage was required then it may be more pragmatic to carry out MLST by published methods. However, for a quick screen enabling the identification of isolates, for instance in the case of an outbreak, then the strategy would seem to be effective and allow the provision of an accurate clonal complex.
ACKNOWLEDGEMENTS
This paper made use of the Campylobacter jejuni Multi Locus Sequence Typing website (http://pubmlst.org/campylobacter/) developed by Keith Jolley and Man-Suen Chan and sited at the University of Oxford (Jolley et al. 2004, BMC Bioinformatics, 5: 86). The development of this site has been funded by the Wellcome Trust.
DECLARATION OF INTEREST
None.





