Background
The protozoan parasite Cryptosporidium can cause disease in both humans and animals. In humans, the disease usually presents with self-limiting watery diarrhoea. However, the infection can be severe in immunocompromised and malnourished individuals [Reference Checkley1]. The incubation period varies between 2 and 10 days, with an average of 7 days [Reference Caccio2]. In the European Union (EU) and European Economic Area (EEA), the yearly notification rate of cryptosporidiosis ranged from 1.67 to 3.26 cases per 100,000 individuals from 2018 to 2022 [3]. In Sweden, laboratory-confirmed cases of cryptosporidiosis are notifiable by law. The yearly notification rate of cryptosporidiosis in Sweden ranged from 5 to 10.5 cases per 100,000 individuals from 2018 to 2022, with a peak in 2019 due to several foodborne outbreaks [Reference Bujila4]. Two large waterborne outbreaks occurred in Sweden in 2010 and 2011 [Reference Bjelkmar5, Reference Widerström6]. Since 2016, most reported outbreaks in Sweden have been foodborne and, to a lesser extent, due to direct contact with infected animals. All reported outbreaks from 2018 to 2022, except for an outbreak in a preschool caused by C. mortiferum, were caused by infection with C. parvum. The most common suspected vehicles in foodborne outbreaks of cryptosporidiosis in Sweden have been green leafy vegetables such as spinach, kale, and arugula [Reference Bujila4]. Several countries in the EU/EEA, as well as the UK, have also reported outbreaks due to the consumption of contaminated food items, such as lettuce mix in Finland in 2008, salad mix in Denmark in 2005, and pre-cut salad leaves in the UK in 2012 [Reference McKerr7, Reference Nasser8].
In the second half of December 2023, an unusually high number of cryptosporidiosis cases were reported to the Swedish notification system for notifiable communicable diseases (Sminet). Out of 21 counties in Sweden, 13 reported cases, and upon request from the Public Health Agency of Sweden (PHAS), Cryptosporidium-confirmed DNA and/or faecal samples were sent to PHAS for molecular typing. A novel subtype family and subtype of Cryptosporidium spp. (later named C. parvum IIγA11) was detected, and a national outbreak was declared and an investigation was initiated to describe the outbreak and identify the source. The outbreak investigation team included members from PHAS, the Swedish Food Agency (SFA), and the affected regional departments of communicable disease control and prevention (CDC). Here, we describe the outbreak of the novel C. parvum IIγA11 and the results of the investigation.
Methods
Descriptive epidemiology
Information on all laboratory-confirmed cryptosporidiosis cases was collected from Sminet, including age, sex (male/female), county, date of disease onset, and sampling date.
Outbreak case definition and case finding
A confirmed case was defined as an individual with laboratory-confirmed cryptosporidiosis notified in Sminet typed to C. parvum IIγA11, domestically acquired infection or unknown travel history, and with the date of disease onset (or date of sampling if missing) between 15 December 2023 and 1 January 2024. A suspected case was an individual with laboratory-confirmed cryptosporidiosis notified in Sminet, with no typing performed, domestically acquired infection or unknown travel history, and with the date of disease onset (or date of sampling if missing) between 15 December 2023 and 1 January 2024.
The outbreak period was defined based on the first confirmed case identified on December 18, which coincided with a rise in notified cases from December 15. The period concluded after the last confirmed case on December 26, incorporating a 7-day incubation period and a noticeable subsequent decrease in the number of reported cases, ending on January 1. After this defined outbreak period, baseline sporadic cases resumed, with two cases detected on January 3 and January 9, which were confirmed to be of different subtypes.
Trawling questionnaire
The national Cryptosporidium trawling questionnaire is web-based and used in routine investigations of notified cases by the regional CDC departments. The results were also available to the outbreak team at PHAS. The questionnaire includes information about symptoms relevant to gastrointestinal illness and possible exposures within 14 days before the onset of disease, such as travel abroad and the consumption of food items including ready-to-eat options like salad bars in grocery stores, where customers can create their own salads by choosing from a variety of greens and other salad ingredients. The questionnaire further includes information about visits to restaurants and cafes, grocery stores for food purchase, contact with pets and farm animals, participation in events, and swimming in recreational water and lakes. The exposure questions are closed-ended questions with three (yes, no, or do not know) or four (yes, probably, probably not, and no) options and with additional open-ended questions about brand and place of purchase or specific type of foods.
Case–control study
A case–control study was undertaken to explore the hypotheses emerging from the trawling questionnaire that certain food items, including specific dishes and ingredients served within salad bars, were the vehicle of transmission. All cases, both confirmed and suspected, who met the outbreak case definitions were included in the study. Controls were recruited from a national random pool of controls (n = 6,586, age > 16 years) available at PHAS. This pool is a statistical selection by the Swedish Statistics Agency to represent Sweden’s population [9]. We included 703 controls, assuming a response rate of 50%, and randomly selected with frequency matching at the group level for age group (16–39, 40–59, and > 60) and sex.
A web-based questionnaire was administered using Survey Generator (Stockholm, Sweden). It included questions about demographics (age, sex, and county), any gastro-enteric symptoms, travelling abroad, and food exposures selected based on the frequency reported in the trawling questionnaires (including consumption of salads and vegetables and eating from self-administered salad bars in grocery stores). From the main provider of salad bars to grocery stores in Sweden, we received a list of dishes included in the salad bars. Some of the salad bar ingredients/dishes were grouped together in the questionnaire, based on similarities either biologically (involving the same ingredients) or practically (being a challenge for responders to visually distinguish between similar dishes). Cases additionally answered questions regarding their symptoms and possible hospitalisation due to the Cryptosporidium infection. The questions were closed-ended with three options for sections regarding clinical symptoms and travel history (yes, no, or do not know) and four options for food exposures (yes, probably, probably not, and no). Time frame was 14 days before disease onset for cases and 14 days before answering the questionnaire for controls. The web-based questionnaire was administered by the regional CDC for cases and by PHAS for controls and was sent out on January 19, 2024.
Data analysis
Controls were excluded if they had experienced symptoms of gastrointestinal illness or travelled abroad 14 days prior to answering the survey. To preserve statistical power, we addressed missing values by substituting them with “no” in questions where no options were selected for respondents who had systematically indicated “yes” or “probably yes” but had not selected “no” or “probably not” in the questionnaire.
This approach assumes that respondents only reported what they had actually consumed and skipped reporting what they did not consume. Options “yes” and “probably” were coded as exposed, and “no” and “probably not” were coded as unexposed. The association between outcome (case/control) and exposures was first assessed through univariable logistic regression. All exposures with an odds ratio (OR) greater than 1 and a P value less than 0.20 were included in the multivariable analysis. To reduce the number of variables, we also decided to include only food items with an exposure rate of at least 40% among the cases.
Two separate logistic models were fitted with case as an outcome, calculating adjusted odds ratios (aOR) and 95% confidence intervals (95% CI). In the first model, we included food exposures including eating from salad bars in grocery shops as one variable (yes/no) as exposures. In the second model, we included the specific ingredients/dishes of the salad bars as exposures. This model only included individuals who had consumed food from these in-store salad bars. The models were adjusted for age (as an integer variable), sex, and county. The model was built using a backward elimination process, keeping those variables/exposures with P value <0.05 using a likelihood ratio test. The presence of confounding was assessed by examining the effect on the coefficient of other variables when adding a variable/exposure to the model. If there was a change of ≥20%, the variable was regarded as a confounder and included in the final model. Exposures that were excluded from the final model were analysed and stratified by exposures remaining in the final model to check for effect modification.
Chi-square tests were used to analyse potential differences in clinical manifestations including fever (> 38 °C), vomiting, diarrhoea, nausea, and abdominal pain, between cases included in the case–control study and Cryptosporidium cases notified in Sminet from 2021 to 2022 who responded to the trawling questionnaire, regardless of typing information.
Statistical analyses were performed using R version 4.3.1 (R Core Team, 2023) in RStudio version 13.1.446 (RStudio Team, 2023).
Molecular and phylogenetic characterisation
A sample selection for molecular characterisation was made at PHAS from the most affected counties and/or priority of samples from cases who had responded to the trawling questionnaire. DNA from faecal samples with sampling date within the outbreak period was extracted using the magLEAD 12gC instrument supplied with magDEA DX MV reagents (Precision System Science Co Ltd., Chiba, Japan). All extractions were performed according to the manufacturer’s instructions. Prior to extraction, oocyst disruption was accomplished by bead beating using a Bullet Blender (Techtum, Sweden). Identification of Cryptosporidium species was done by amplification of the small subunit rRNA (ssu rRNA) gene by PCR followed by bi-directional sequencing of the PCR products [Reference Xiao10, Reference Xiao11]. Subtyping was done by amplification of the 60 kDa glycoprotein (gp60) gene followed by bi-directional sequencing of the PCR products using primers described by Alves et al. [Reference Alves12]. Editing and analysis of sequences were done using CLC Main Workbench (Qiagen, Aarhus, Denmark, version 21).
A phylogenetic tree was constructed using C. parvum reference sequences from foodborne outbreaks linked to green leafy vegetables in Sweden as well as known Cryptosporidium species and subtypes detected in Sweden retrieved from the NCBI GenBank database, and analysis was performed on the novel gp60 DNA sequence generated in this investigation. A phylogenetic tree was generated using the neighbour-joining method based on Kimura’s 2-parameter model [Reference Kumar13]. To estimate robustness, bootstrap proportions were computed after 1000 replications. All analyses were conducted in MEGA XI (accessed on 9 February 2024).
Trace-back investigation
In Sweden, there is one major producer of these salad bars. In collaboration with the competent authority of the salad bar supplier, SFA carried out a trace-back investigation of the implicated products based on the results of the case–control study.
Results
Descriptive epidemiology
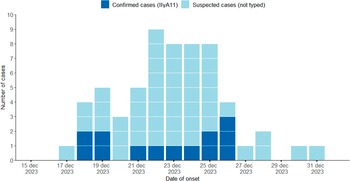
In total, 60 cases were identified as outbreak cases, of which 13 were confirmed and 47 suspected. The median age was 44 (range 16–81 years), 73% were women (Figure 1). The majority of cases (61%) were women between 20 and 59 years. Among the 21 counties in Sweden, 13 notified cases, with the highest numbers in two counties located in the southern region of Sweden (Jönköping 7.3 and Halland 5.3 cases per 100,000 inhabitants). Confirmed cases were detected in six counties. The onset date of disease ranged from 17 to 31 December 2024, with a peak observed between 22 and 25 December (Figure 2).

Figure 1. Number of suspected (n=47)and confirmed (n=13) Cryptosporidium outbreak cases, Figure 3by age group and sex in Sweden between 15 December 2023 and 1 January 2024 (n=60).

Figure 2. Number of suspected (n=47) and confirmed (n=13) Cryptosporidium outbreak cases by date of disease onset in Sweden between 15 December 2023 and 1 January 2024.
Clinical manifestations
The most frequent symptoms reported among cases in the study questionnaire (n = 34) were diarrhoea (100%), nausea (88%), and abdominal pain (79%) with no significant differences compared to historical cases from 2021 and 2022 (n = 287). However, fever (76%; P < 0.01) and vomiting (56%; P < 0.01) were more common among cases with C. parvum IIγA11 compared to historical cases. The hospitalisation rate reported among cases with C. parvum IIγA11 was 32%, with a median age of 32 years (historical data not available).
Trawling questionnaire
The trawling questionnaire had 38 responders, and the result showed that the majority of cases had eaten from salad bars in grocery stores (70%), as well as the following food items: tomato (82%), cucumber (81%), onion (61%), minced beef (55%), iceberg lettuce (53%), and spinach (50%). Exposure to kale was not included in the trawling questionnaire.
Case–control study
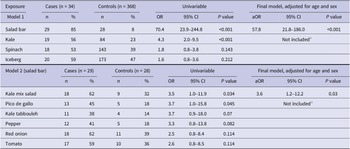
In total, 34 cases (8 confirmed and 26 suspected) and 422 controls completed the study questionnaire, resulting in response rates of 57% and 60%, respectively. Fifty-four controls were excluded. As a result, 368 controls were included in the analysis. Characteristics of cases and controls are presented in Table 1.
Table 1. Characteristics of cases and controls in a case–control study of the Cryptosporidium outbreak in Sweden between 15 December 2023 and 1 January 2024

Note: Number and percentage of cases (n = 34) and controls (n = 368) per age group, sex, and county.
The results of the univariate analysis are presented in Supplementary Table 1. The results of the first model showed that cases were more likely to have consumed food from grocery store salad bars compared to controls (aOR: 58, 95% CI: 22–186) (Table 2). For items included in the first model, it was not possible to determine from the questionnaire whether food items such as kale, spinach, and iceberg lettuce were purchased from a store shelf, consumed from the salad bar, or elsewhere. The second model, which examined the consumption of specific dishes from the grocery store salad bar, showed that cases were more likely to have consumed kale mix salad compared to controls (aOR: 3.6, 95% CI: 1.2–12.2).
Table 2. Results from univariable and multivariable logistic regression models in a case–control study of the Cryptosporidium outbreak in Sweden between 15 December 2023 and 1 January 2024.

Note: Number and percentage exposed, odds ratio (ORs) from univariable logistic regression models and adjusted OR (aOR) from multivariable logistic regression models adjusted for age and sex.
a Not significant when fitting the final model.
Molecular characterisation and phylogenetic analysis
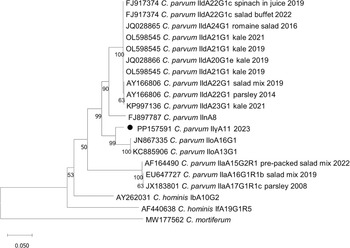
A total of 20 samples were selected for molecular typing, out of which 13 samples were of the novel subtype family and subtype C. parvum IIγA11 (confirmed cases). Remaining seven samples were C. parvum subtypes IIaA15G2R1 (n = 1), IIaA17G1R1c_variant (n = 1), and IIdA20G1e (n = 4). For one sample, the subtyping was unsuccessful. These samples (n = 7) were excluded from the analysis. Representative nucleotide sequences, gp60 and ssu rRNA, of the novel subtype family and subtype C. parvum IIγA11 were submitted to the NCBI GenBank database under the following accession number PP157591 for gp60 and PP328976 for ssu rRNA. The novel gp60 sequence did not cluster with any of the previously reported subtypes of foodborne outbreaks where green leafy vegetables were the suspected source of contamination. Instead, the closest sequence matches were C. parvum IIoA16G1 and IIoA13G1 (Figure 3).

Figure 3. Phylogenetic relationships between C. parvum IIγA11 from the Cryptosporidium outbreak in Sweden between 15 December 2023 and 1 January 2024 (filled circle) andsequences from foodborne outbreaks linked to green leafy vegetables in Sweden, known Cryptosporidium spp. and subtypes detected in Sweden and closest sequence match to C. parvum IIγA11 retrieved from the NCBI GenBank database.
No batches of food items were left to be analysed when the potential source of the outbreak was identified.
Trace-back investigation
Based on the results of the case–control study, the trace-back investigation focused on kale in kale mixed salad from salad bars. One food processing company in Sweden delivered all salad products, including kale mixed salad, to salad bars in grocery stores. This company received kale several times a week, with total quantities of about three to four tons per week. During November and the beginning of December 2023, kale was almost exclusively produced by one single grower in Sweden. However, from December 11, there was a shift to deliveries from other growers in Sweden, Spain, and Belgium. It was not possible to link kale from any specific grower as the likely source of the outbreak.
Outbreak control measures
PHAS posted information regarding the outbreak on their website on 18 January 2024, providing weekly updates until the outbreak was officially declared over on 16 February 2023. The company responsible for delivering salad bars to grocery stores was contacted by SFA and recommended to investigate the routine measures for washing vegetables. An urgent inquiry for cases in other European countries was posted on EpiPulse on 19 January 2024 (2024-FWD-00003). None of the five countries who responded to the inquiry had experienced an increase in the notification rate of cryptosporidiosis.
Discussion
The national outbreak of cryptosporidiosis occurred in December 2023 in Sweden and involved 60 cases, suspected and confirmed, which were largely concentrated in two counties in the south. The peak (shape) of the epidemic curve indicated a fresh produce with short shelf life as a potential vehicle of the outbreak. The trawling questionnaire indicated a vegetable source, which also was the finding in the case–control study where the results showed that consumption of food items from salad bars in grocery stores could account for 85% of the cases, with kale mix salad within the salad bar dishes explaining 62% of cases. One limitation could be the misclassification of “yes” as “no” due to replacing missing answers with “no,” potentially leading to an underestimation of the effects of the exposures. However, there were no missing answers for the question regarding the exposure consumption from salad bars.
The outbreak was most likely caused by kale mixed salad from salad bars in grocery stores. Since 2018, 17 outbreaks have been reported in Sweden, and in 65%, green leafy vegetables, especially kale but also arugula and salad mixes, were the suspected vehicle of infection [Reference Bujila4]. No outbreaks due to contaminated drinking water or recreational water have been reported in Sweden since 2016 [Reference Bujila4].
We detected a novel subtype family and subtype, C. parvum IIγA11. The subtype IIoA16G1, one of the two closest sequence matches, was first identified in a Swedish human case who had travelled to Thailand and it has additionally been identified in four cases of HIV-infected patients in Thailand [Reference Insulander14, Reference Sannella15]. The other closest sequence match IIoA13G1 has been identified in one human case in New Zealand in 2019 [Reference Garcia16]. The subtype family IIo has also been detected in bamboo rats (Rhizomys sinensis) in China [Reference Liu17]. Clinical manifestations of this outbreak differed in some respects from previously notified cryptosporidiosis cases. Infection with this novel subtype exhibited a higher frequency of fever and vomiting. The hospitalisation rate of cases in this outbreak, as reported in the questionnaire, was higher (32%) compared to a previous study on cryptosporidiosis cases in Sweden from 2006 to 2008, which reported a hospitalisation rate of 15%, regardless of species/subtype and travel history [Reference Insulander14]. Identification of new species and subtypes of Cryptosporidium has accelerated in recent years and knowledge of the mechanisms behind the emergence of new subtypes with potential public health impact is critical for understanding cryptosporidiosis epidemiology and for outbreak investigations [Reference Feng, Ryan and Xiao18].
Foodborne outbreaks of cryptosporidiosis are notoriously difficult to investigate due to the long incubation period of the infection and thus often no food items are left in stores to analyse. This is not uncommon in outbreaks where green leafy vegetables are the suspected source since they are freshly produced or quickly consumed or discarded and thus limiting the possibility of sampling and analysis. Further, if sampling is possible, analysis of Cryptosporidium is challenging in matrices including vegetables with few existing standardised protocols, although there is a microscope-based ISO method for analysing leafy greens [19]. Concerning food safety, green leafy vegetables are a particular challenge since they are served raw, and as such, microbiological hazards are significant [Reference Mogren20].
Outbreak investigations of cryptosporidiosis are likely to be subject to recall bias, partly due to the long incubation period, which could lead to misclassification of exposure as well as posing difficulties in source tracing. Recall bias is also likely to be more pronounced when asking about food consumed in buffets due to the extensive variety of dishes typically available. It is plausible that respondent’s answers reflect their usual selections from salad buffets rather than precise recollections of the timing associated with their illness. Additionally, the timing of the questionnaire administration is a potential bias. Both cases and controls were given the questionnaire in mid-January, but cases were asked to recall their food intake from 2 weeks before disease onset, which corresponded to mid-December, while controls reported on their food intake for the last 2 weeks before answering the questionnaire. We considered asking controls about their food intake during the same period as the cases to minimise this time discrepancy. However, we determined that asking controls about a more recent period would likely yield more accurate information. This decision was made with the understanding that salad buffet choices are generally habitual, reflecting typical consumption patterns, and thus less likely to be significantly influenced by seasonal events such as Christmas or end-of-semester activities.
Kale was identified as the salad bar item associated with cases. As previously mentioned, kale has been linked to past outbreaks of cryptosporidiosis, possibly attributed to its cultivation above soil level and challenges in thorough washing. Raw green leafy vegetables, especially kale, may pose a health risk to consumers, and as such, the prevention of contamination should be emphasised throughout the production chain.
In general, there is a lack of widespread and systematic genotyping to detect species and link cases, as well as a lack of suitable methods for testing food items and thus linking them to cases. This lack of resources and methodologies hampers proper investigations and public health interventions in foodborne outbreaks of cryptosporidiosis.
Conclusion
Our multidisciplinary investigation suggests that salad bars in grocery stores and particularly kale mix salad from the salad bars were the sources of this outbreak of a novel subtype family and subtype, i.e. C. parvum IIγA11. This new subtype with potential different clinical manifestations should continue to be monitored. We did not obtain microbiological confirmation from food items due to the short shelf life and the long incubation period of cryptosporidiosis. Kale and other green leafy vegetables are often consumed raw, which poses challenges for ensuring food safety throughout the production chain. This investigation, along with previous ones where kale was the suspected source of contamination, indicates that despite safety procedures in the production chain, Cryptosporidium may still contaminate these products leading to outbreaks and posing a public health risk.
Supplementary material
The supplementary material for this article can be found at http://doi.org/10.1017/S0950268824001432.
Data availability statement
Ssu rRNA and gp60 sequence data from representative samples of C. parvum IIγA11 are available at the NCBI GenBank database with accession number PP328976 and PP157591, respectively.
Acknowledgements
We would like to acknowledge the county medical officers in the following counties: Blekinge, Dalarna, Gävleborg, Halland, Kronoberg, Skåne, Stockholm, Södermanland, Uppsala, Västerbotten, Västra Götaland, and Östergötland for their valuable contribution to the outbreak investigation. We would also like to acknowledge all the control group participants from Hälsorapport and cases that took the time to fulfil the questionnaire; the clinical microbiological laboratories for providing isolates; Marie Rapp, Fredrik Garli, and Kate Lillepold (at PHAS) for help with Hälsorapport; Leigh Davidsson for contribution in the outbreak investigation; and Nasanin Hashemi for aid in statistical analysis. Further, we would also like to acknowledge Maria Reinius for help with the case–control study survey. We would also like to thank Professor Lihua Xiao at South China Agricultural University for his great contribution to typing discussions and analysis.
Authors contribution
IB, AO, AH, LA, CR, and MR were part of the outbreak team. IB, AO, MR, and IG contributed to the design and implementation of the case–control study. AH coordinated the investigation at the national level. AO, SKB, IB, IG, and MR were responsible for data analysis and interpretation of the case–control study. LA and IB coordinated and performed the laboratory investigations. IB and MarL preformed the microbiological analysis. IH and AMG were responsible for the outbreak investigation in their respective regional CDC. MatL was responsible for the trace-back investigation. IB and AO coordinated, drafted, and finalised the manuscript. All authors contributed to the revision of the draft manuscript and approved the final version.
Funding statement
The authors are fellows of the ECDC Fellowship Programme, supported financially by the European Centre for Disease Prevention and Control. The views and opinions expressed herein do not state or reflect those of ECDC. ECDC is not responsible for the data and information collation and analysis and cannot be held liable for conclusions or opinions drawn.
Competing interest
The authors declare no competing interests exist.
Ethical statement
The data used for the outbreak investigation were collected as part of PHAS mandate for infectious disease surveillance and control as defined by national legislation. Hence, ethical approval was not necessary.