Global prospective surveillance data suggest a recent re-emergence of Mycoplasma pneumoniae (M. pneumonia) infection in Europe and Asia. This delayed re-emergence is striking because it occurred sometime after non-pharmaceutical interventions against COVID-19 were discontinued, and a possible circulation of M. pneumoniae is usually described as endemic, punctuated with cyclic epidemics every 3 to 5 years [Reference Beeton1]. M. pneumoniae beyond the COVID-19 pandemic surveillance is needed [Reference Meyer Sauteur2, Reference Meyer Sauteur3].
In our paper, we have highlighted an outbreak of M. pneumoniae infection in Nord Franche-Comté Hospital, a French district general hospital with a capacity of 1,216 beds and about 100,000 visits to its emergency rooms per year [Reference Zayet4], so as to alert the medical community about a possible emergence of this pathogen and what appears to be its changing attributes. Our patients reside in Franche-Comté (Belfort-Montbeliard), a region of about 300,000 habitants in the East of France, close to the Swiss and German borders. In the period from 14 November 2022 to 31 January 2023 (the same period in the previous year), only one patient (a 15-year-old man) was diagnosed in our hospital for M. pneumoniae-associated community-acquired pneumonia (CAP) (Figure 1). Our series consisted of 13 confirmed cases of M. pneumoniae CAP (11 adults and 2 older children) in hospitalized patients in Nord Franche-Comté Hospital between 14 November 2023 and 31 January 2024. In adult inpatients, the mean age was 45.5 years (range: 19–70) with a male predominance (72.7%). All patients were considered as immunocompetent mostly without comorbidities (Charlson comorbidity index mean at 0.5 (range: 0–2)). Tobacco consumption was noted in 45.5% of adult patients (n = 5). The 13 patients had no direct contact with each other, and no significant epidemiological links were discovered. Two adult patients reported similar respiratory presentations in their children. M. pneumoniae was a part of a polymicrobial infection in only one patient, and a second one had co-infection with confirmed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (paucisymptomatic form with no respiratory symptoms initially). Diagnosis in all patients was based on positive serology (IgM and IgG) and/or positive specific or multiplex reverse transcription polymerase chain reaction (RT-PCR) on respiratory specimens (nasopharyngeal swab or bronchoalveolar lavage). Pulmonary consolidation or diffuse infiltrates were described in our patients with bilateral patterns in 72.7% of cases (n = 8/11). All adult patients were treated with antimicrobial drugs (spiramycin (n = 9), azithromycin (n = 1), and levofloxacin (n = 1)) with a favourable outcome. Two patients were transferred to the intensive care unit (ICU) but were not mechanically ventilated. All patients required oxygen supplementation with a mean duration of oxygen support of 6.2 days (range: 1–13) (Table 1).
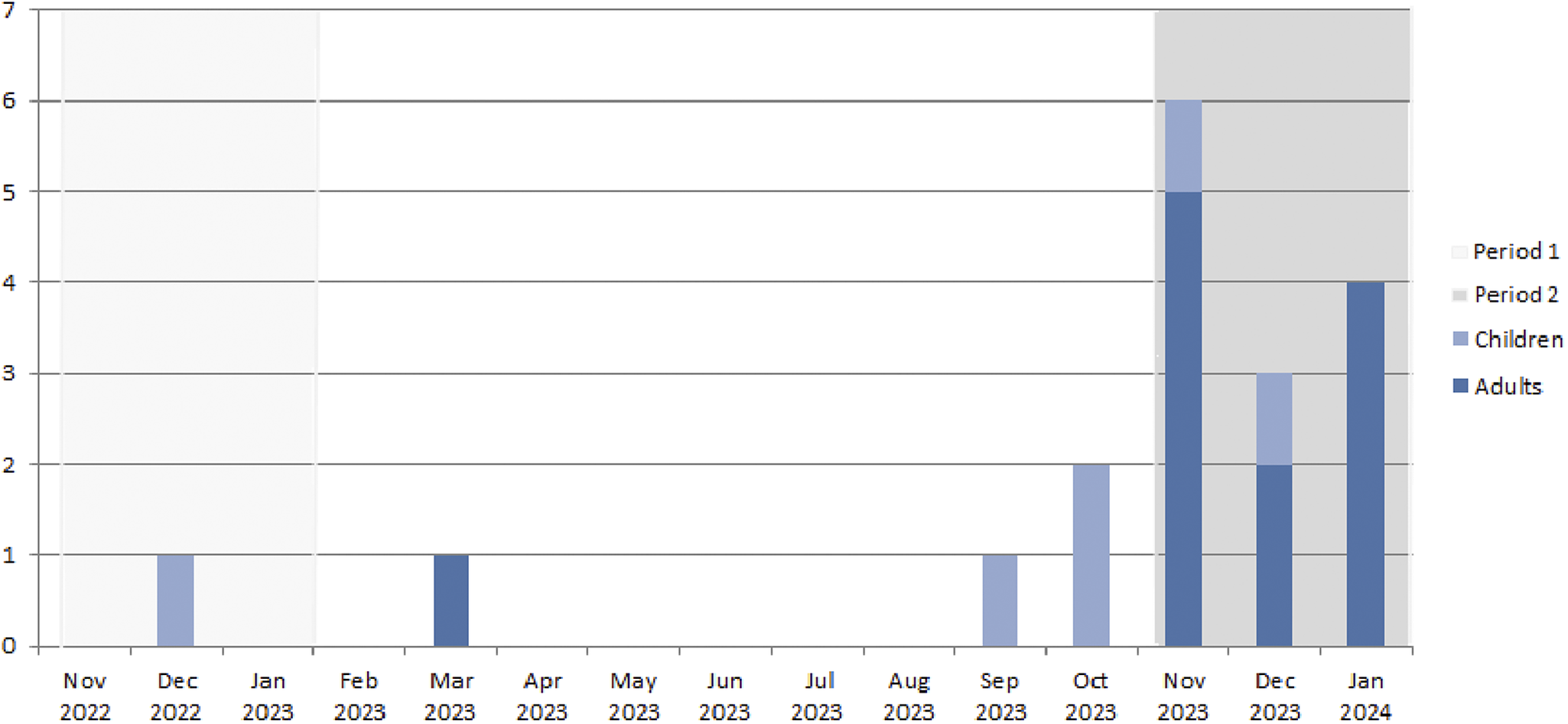
Figure 1. Monthly distribution of Mycoplasma pneumoniae infections (confirmed on positive IgM or positive RT-PCR on respiratory specimen) in children and adults in Nord Franche-Comté Hospital, 14 November 2023–31 January 2024 (Period 1: 14 November 2022 to 31 January 2023, Period 2: 14 November 2023 to 31 January 2024).
Table 1. Characteristics of patients hospitalized for Mycoplasma pneumoniae infections in Nord Franche-Comté Hospital, 14 November 2023–14 January 2024
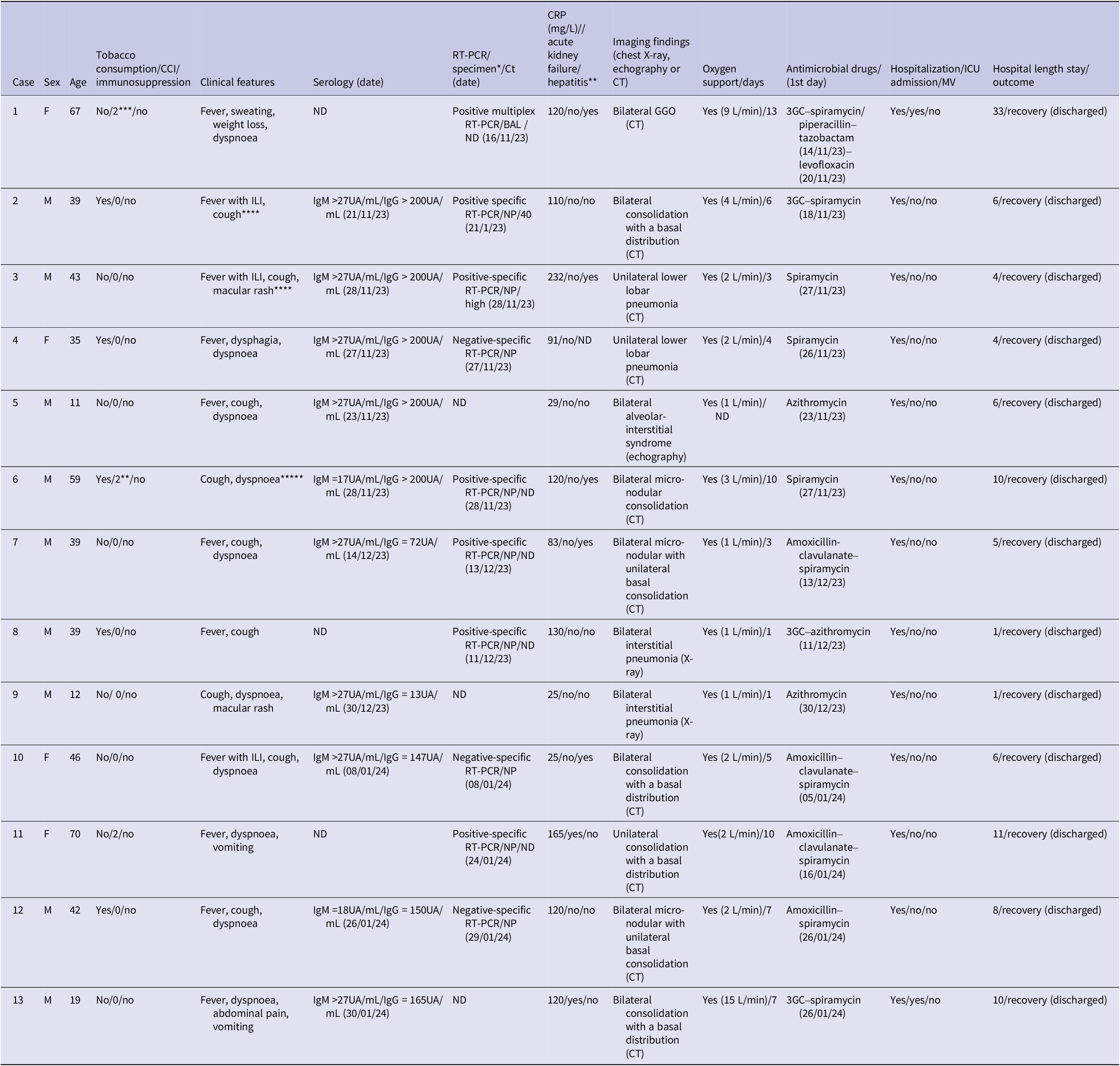
Abbreviations: 3GC: third-generation cephalosporin; BAL: bronchoalveolar lavage; CCI: Charlson Comorbidity Index; CT: computed tomography; F: female; ILI: influenza-like illness; GGO: ground-glass opacities; M: male; ND: not determined/done; NP: nasopharyngeal swab; MV: mechanic ventilation; RT-PCR: reverse transcription polymerase chain reaction; ICU: intensive care unit.
*We identified a polymicrobial infection in patient 1 with associated other bacterial species (Serratia marcescens and Mycobacterium tuberculosis). Patient 7 was initially hospitalized for confirmed SARS-CoV-2 infection with concomitant Mycoplasma pneumoniae community-acquired pneumonia.**Five patients (1, 6, 7, 10, 11, and 13) presented hepatitis with cholestasis. ***Three patients (1, 6, and 11) with Chalson Comorbidity Index at 2 had underlying cardiovascular diseases. ****Patients 2 and 3 reported similar respiratory presentations in their children; *****Patient 6 reported a recent travel in Guyana since 2 weeks.
We performed an NP swab for specific RT-PCR QIAstat® for M. pneumoniae by sequence analysis of the p1 adhesin gene. We also performed LBA specimens for DiagCORE®, Multiplex Respiratory Panel 2 – SAT Dx., which detects viral and bacterial pathogens including human mastadenovirus A–G (formerly adenovirus), primate bocaparvovirus 1 + 2 (formerly bocavirus), human (hMPV), rhinovirus/enterovirus, influenza A virus (as no subtype, subtype H1, H1N1/2009, or H3), influenza B virus, human respirovirus 1 or 3, human orthorubulavirus 2 or 4 (formerly human parainfluenza virus types 1–4), human orthopneumovirus, M. pneumoniae, Legionella pneumophilia, Bordetella pertussis, and Chlamydia pneumoniae, coronavirus (differentiating HKU1, NL63, OC43, or 229E), human metapneumovirus A/B, and SARS-CoV-2.
Over a two-and-a-half-month period from mid-November 2023 to end-January 2024 onwards, we observed an increased number of hospitalized patients diagnosed with pneumonia caused by M. pneumoniae, in Nord Franche-Comté Hospital, in the East of France. In our analysis, we preferred to exclude the two children because it is not unusual for children to be hospitalized for M. pneumoniae infection during the cold period. Larcher et al. have also reported an increased incidence of M. pneumoniae CAP in young adults hospitalized in a French university hospital (n = 6) during the first three weeks of November, with similar findings in patients’ characteristics [Reference Larcher5]. This case series suggests that the pathogenicity of M. pneumoniae, even in adult immunocompetent patients without comorbidities, has changed. All patients presented mild-to-severe pneumonia requiring supplemental oxygen support, with a high-flow nasal cannula in the oldest patient. National data from epidemiologic surveillance and the renal network of hospital laboratories showed that the number of PCR detections of M. pneumoniae increased significantly from October 2023 onwards; this number tripled between weeks 40 and 46 in 2023, with a further increase in week 47, especially in the under-15 and 15–44 age groups [6]. M. pneumoniae presented as a co-infection associated with a second pathogen (Mycobacterium tuberculosis and SARS-CoV-2) in two patients, suggesting that it may preferentially concern patients with pre-existing lung parenchymal involvement/disease. The highest macrolide resistance M. pneumoniae (MRMp) rate was detected in Taiwan from April 2018 to March 2019 with 42 of 53 isolates (79%) [Reference Meyer Sauteur7], with a high prevalence also in Japan [Reference Meyer Sauteur7, Reference Tashiro8]. In France, the MRMp detection rate was 13% (n = 3) and 12% (n = 2) in 2018 and 2019, respectively; however, no resistant strains were found in 2017 and 2020 [Reference Meyer Sauteur7]. In our series, only one patient did not respond to macrolides after 5 days of treatment with ICU admission, raising the issue of a possible MRMp (no antimicrobial susceptibility testing was performed for this patient, which limits this hypothesis). One final point to emphasize is the discordance between serology and PCR results (which were negative in three patients and low positive in two patients). This can be explained by the fact that the respiratory sample was performed 1–3 days after the introduction of antibiotics in these patients.
One of the limitations of our paper was the absence of baseline number of cases in our region pre-COVID-19. This does not lead us to conclude for certain if these cases represent a truly unusual increase or simply a re-emergence of M. pneumoniae following the COVID-19 pandemic. Nevertheless, providers should more seriously consider M. pneumoniae in their differential diagnosis and testing choices; M. pneumoniae serology still has a place in molecular biology and should be performed only in patients with negative PCR and compatible symptoms without other diagnoses. Rapid PCR negativity seems to be good evidence that patients are non-contagious shortly after the start of antimicrobial drugs.
We recommend that worldwide surveillance of M. pneumoniae is needed [Reference Meyer Sauteur2, Reference Meyer Sauteur3] at the present time.
Data availability statement
Data are available on request due to privacy restrictions. The data presented in this case study are available on request from the corresponding author.
Author contribution
Conceptualization: C-E.L., J.P., T.K., V.G., S.Z.; Data curation: C-E.L., S.P., S.Z.; Funding acquisition: C-E.L., J.P.; Investigation: A.H., S.P., T.K., Y.M., S.Z.; Resources: A.H., S.P., Y.M., S.Z.; Software: A.H., S.P., Y.M.; Methodology: J.P., T.K., S.Z.; Formal analysis: S.P., S.Z.; Supervision: T.K.; Validation: T.K., V.G.; Writing – review & editing: T.K., V.G.; Visualization: V.G.; Writing – original draft: S.Z.
Funding statement
This research received no external funding.
Informed consent statement
We made sure to keep participant data confidential and in compliance with the Declaration of Helsinki.
Competing interest
The authors declare no conflicts of interest.





