INTRODUCTION
The genus Salmonella includes more than 2400 different known serovars [Reference Nair1, Reference Martinez-Urtaza2]. Salmonella serovars are associated with considerable morbidity and mortality among livestock, thereby posing a significant threat to animal health and well-being and, as a result, to human health [Reference Martinez-Urtaza2–Reference Gorman and Adley4].
Non-typhoidal Salmonella serovars are increasing in importance as significant pathogens of both human and animals. According to the World Health Organization, there are about 17 million cases annually of acute gastroenteritis or diarrhoea due to non-typhoidal salmonellosis, with 3 million deaths [Reference Rabsch, Tschape and Baumler5, Reference Ling and Wang6].
Phenotypic methods play an important role in identification to genus level. Serotyping, based on the Kauffmann–White scheme, remains the standard for classification of Salmonella isolates in outbreak investigations but now has been supplemented by a range of molecular genotyping methods [Reference Botteldoorn7–Reference Johnson13].
In recent years, molecular-based techniques, such as plasmid profile analysis, ribotyping, random amplified polymorphic DNA analysis, and pulsed-field gel electrophoresis (PFGE) have been shown to be useful methods for discrimination among isolates of Salmonella spp. Among these techniques, PFGE is currently considered to be one of the most reliable typing procedures [Reference Tsen and Lin14–Reference Garaizar17].
In Tunisia, the surveillance for Salmonella enterica is carried out by the National Centre of Enteropathogenic Bacteria (Salmonella, Shigella, and Vibrio cholerae). Annually, about 2000 Salmonella strains are reported from all over Tunisia to the National Centre for Enteropathogenic Bacteria. Any Salmonella strains isolated are analysed and serotyped.
Identification of isolates has been limited to the serovar level, without any additional characterization. Molecular typing data are useful for epidemiological studies; this information would facilitate the identification of the clonality of isolates and, in turn, would determine the epidemiological relationship and prevalence of different strains of Salmonella [Reference Lindqvist, Siitonen and Pelkonen18–Reference Nair23]. Molecular techniques were first introduced for Salmonella analysis in 2001. According to statistical data of an 11-year study period from 1994 to 2004 (R. Ben Aissa, unpublished observations), three Salmonella serovars were the most commonly isolated in Tunisia: Salmonella enterica serovars Enteritidis, Anatum, and Corvallis. Typhimurium is also of importance in Tunisia since it showed obvious peaks in different categories (food, human, animal and environment) over the 11-year study period and was therefore also included in our study.
The objectives of this study were to determine the extent of genetic variation and clonality among food and clinical strains of S. Enteritidis, S. Anatum, S. Corvallis, and S. Typhimurium in Tunisia. Specifically, this has been achieved by examination of susceptibility to common antibiotics, plasmid analysis, and PFGE patterns.
METHODS
Bacterial strains
A total of 72 Salmonella strains from different sources (mainly food and human stool samples) were isolated and serotyped during the period 2001–2004: Anatum (40), Enteriditis (18), Corvallis (8), and Typhimurium (6).
Serogrouping and serotyping
Salmonella strains were positively identified and serotyped according to the Kauffmann–White scheme with the use of antiserum (Bio-Rad, Marnes-la-Coquette, France). Serogrouping and serotyping were performed by slide agglutination to identify the somatic O antigen and flagellar H antigens.
Antimicrobial susceptibility
Antimicrobial susceptibility testing was done using standard methods (disc diffusion method) using Mueller–Hinton agar, and interpreted according to the antibiogram guidelines of the French Committee of Microbiology (Société française de Microbiologie, 2002). Antimicrobials used for testing were ampicillin (AMP) 10 μg; cephalothin (CEF) 30 μg; ticarcillin (TIC) 75 μg; cefotaxime (CTX) 30 μg; chloramphenicol (CHL) 30 μg; amoxicillin (AMX) 25 μg; cefoxitin (FOX) 30 μg; amoxicillin+clavularic acid (AMC) 20/10 μg; trimethoprim–sulfamethoxazole (SXT) 1·25/23·7 μg; gentamicin (GEN) 10 μg; nalidixic acid (NAL) 30 μg; sulfonamide (SSS) 200 μg; tetracycline (TET) 30 μg; kanamycin (KAN) 30 IU; ciprofloxacin (CIP) 5 μg; streptomycin (STR) 10 IU; and ofloxacin (OFX) 5 μg. S. Choleraesuis strain ATCC14028 strain was used as a control. Characterization of strains as susceptible, intermediately resistant, or resistant was determined by Osiris software version 3.x (Bio-Rad).
Plasmid analysis
Plasmid DNA was isolated by the alkaline lysis method as described previously [Reference Vassu24]. Plasmids were sized in comparison to E. coli V517 strain and compared by the use of Bionumerics software (Applied Maths, Kortrijk, Belgium). The molecular mass of the plasmids was calculated by comparison with plasmids in V517 and images normalized accordingly.
Genomic fingerprinting by PFGE
The XbaI PFGE patterns were determined for all 72 Salmonella strains using previously described PFGE methods with modifications, as described previously [Reference Pasmans25]. PFGE was performed on a 1% agarose gel (Bio-Rad) using CHEF DR III apparatus (Bio-Rad) in 0·5× TBE (Tris–borate–EDTA) buffer at 14°C with 6 V/cm at a field angle of 120°: block 1, 8·5 h, with initial switching time of 7 s to final switching time of 12 s; block 2, 10·5 h, with initial switching time of 20 s to final switching time of 40 s. Gels were stained with ethidium bromide and photographed. A lambda DNA ladder with size range of 48·5 kb to 1 Mb (Amersham Biosciences, Buckinghamshire, UK) was used as a DNA size standard.
Numerical analysis of PFGE profiles
Together with visual analysis of the PFGE profile, a numerical analysis after conversion, normalization, and analysis of similarity in band pattern was performed using MVPS3.31 software (Media Cybernetics, GA, USA). Similarities between profiles were calculating using Dice coefficient, with a maximum position tolerance of 1%. PFGE patterns obtained were clustered by UPGMA. The capital letter A, E, C, and T were used to designate the different serovars: Anatum, Enteritidis, Corvallis, and Typhimurium. The numerical suffix between brackets was used to designate the different years of isolation.
RESULTS
Antimicrobial susceptibility
Of the 72 Salmonella isolates, 62·5% (27/72) were resistant to one or more antimicrobials (Table 1). Twelve (2 clinical and 10 food isolates) of the 40 S. Anatum isolates were sensitive to all antibiotics tested. One clinical strain was drug multiresistant with resistance to 11 different antibiotics (Table 1).
Table 1. Antibiotic resistance profile

AMC, Amoxicillin+clavularic acid; AMP, ampicillin; AMX, amoxicillin; CAZ, ceftazidime; CEF, cephalothin; CHL, chloramphenicol; CIP, ciprofloxacin; CTX, cefotaxime; FOX, cefoxitin; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; OFX, ofloxacin; SSS, sulfonamide; STR, streptomycin; SXT, trimethoprim–sulfamethoxazole; TET, tetracycline; TIC, ticarcillin.
No great variation of susceptibility among the 18 strains of S. Enteritidis was observed (Table 1). Seven strains from different food sources were resistant to tetracyclines, and one from turkey meat to streptomycin. Among the eight strains of S. Corvallis, only strains isolated from turkey meat were resistant. Of the six S. Typhimurium, only one isolate from white cheese was multi-resistant (eight antibiotics: AMP, STR, TET, CHL, AMX, AMC, TIC, and SSS). This strain was phage-typed at the French National Center for Salmonella (Institut Pasteur, Paris, France) by the method described by Anderson et al. [Reference Anderson26] and was defined as multi-resistant serovar Typhimurium DT104.
Plasmid profiles
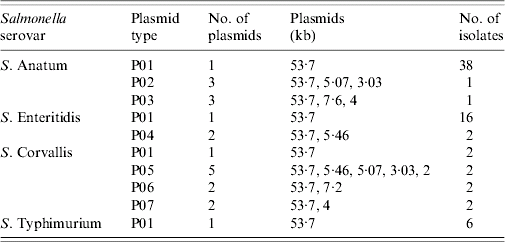
Seven different plasmid profiles with 1–5 plasmids were identified (Table 2). The most prevalent plasmid profile was P01, containing one plasmid of 53·7 kb; PFGE in 62 out of 72 isolates (86%) exhibited this.
Table 2. Number of plasmids and plasmid size of each plasmid profile in the 72 Salmonella strains characterized in this study
PFGE
Four out of 72 isolates were untypable because of DNA degradation. The PFGE patterns of XbaI-digested chromosomal DNA of the remaining 68 isolates are summarized in Table 3.
(i) S. Anatum. A total of 11 PFGE patterns were identified (Fig. 1a). A summary of these patterns is shown in Table 3. Most of these strains belonged to clusters X01 (10/40 isolates) and X02 (9/40 isolates). Cluster X01 was detected in strains isolated from food and human stool samples during the years 2002 and 2003 in the same seasons. The PFGE patterns of strains belonged to cluster X01 were 100% genetically similar (identical profiles). Cluster X02 was detected in food strains during different seasons of the year 2001. The genetic similarity among X01 and X02 was 80%. The remaining strains clustered in one of nine different PFGE patterns. Seven of these clusters (X03, X04, X06, X08, X09, X10, X11) were unique for food isolates.
(ii) S. Enteriditis. PFGE permitted the resolution of XbaI macrorestriction fragments of the 18 S. Enteritidis isolates into three distinct clusters (Table 3). The largest cluster was X01 (14/18 isolates). All 14 isolates were produced between 2001 and 2002 throughout different seasons. The genetic similarity among these three PFGE clusters was 64% (Fig. 1b).
(iii) S. Corvallis. The eight strains of S. Corvallis were assigned to four different clusters (Table 3). All food strains of 2001 clustered in pattern X0A, while human stool strains of 2001 clustered alone in pattern X0C (unique profile) with 62% similarity with pattern X0A (Fig. 1c). The remaining PFGE patterns are summarized in Table 3.
(iv) S. Typhimurium. PFGE patterns of XbaI-digested chromosomal DNA of six S. Typhimurium isolates are shown in Fig. 2. Three patterns were observed: X0A (three strains), X0B (2 strains) and X0C (one strain) (Table 3).
Table 3. The PFGE patterns and plasmid types of the 68 Salmonella enterica serovars: Anatum, Enteriditis, Corvallis, and Typhimurium from patients, food and environmental samples in Tunisia

* Diarrhoeagenic cases.

Fig. 1. Dendogram showing percent similarity calculated by the Dice similarity index of PFGE restriction endonuclease digestion profiles among the 66 Salmonella isolates: (a), S. Anatum; (b), S. Enteritidis; (c), S. Corvallis. The different patterns, sources, year of isolation and number of strains are indicated.

Fig. 2. PFGE patterns of XbaI digests of chromosomal DNA of S. Typhimurium. The numerical prefix was used to designate the serovar of the strain. The alphabetical suffix was to designate the code of the strain in our laboratory. Salmonella enterica serovar cholereasuis ATCC14028 strain was used as a control. M, lambda DNA ladder; lane 1, S. Choleraesuis ATCC14028; lane 2, ST21(01); lane 3, ST1031(01); lane 4, ST2157(01); lane 5, ST1161(02); lane 6, ST179(03); lane 7, ST299(03).
DISCUSSION
This study has utilized a combination of phenotypic and genotypic typing methods to define relationships between 72 strains of S. Anatum, S. Enteritidis, S. Corvallis, and S. Typhimurium from human stool, food, animal, and environmental sources in Tunis. In this investigation 62·5% of the 72 isolates of the four serovars were resistant to at least one antibiotic (tetracycline), and most were multiresistant (to tetracyclines, streptomycin, and chloramphenicol). Both resistance and multiresistance were more common in S. Anatum, S. Corvallis and S. Typhimurium. One S. Anatum strain, from a human stool diarrhoeagenic case, was resistant to 11 antibiotics. In contrast, isolates of S. Enteritidis were predominantly drug-sensitive.
S. Typhimurium DT104 with multidrug resistance is an important international human pathogen, and it is widespread in Western and Eastern Europe, North America, and the Middle East [Reference Martinez-Urtaza2, Reference Humphrey27]. In our study, one S. Typhimurium strain, from a dairy product (cheese), was multidrug-resistant type DT104 (according to the phage typing). This strain was resistant to eight antibiotics.
When studied by PFGE, within S. Anatum the majority of strains fell into two major PFGE patterns (X01, X02). Strains with PFGE patterns of X01 were detected exclusively in the winter months in Tunis in 2002 and 2003, and were isolated from different sources. Isolates of the X02 pattern were detected throughout 2001 from different food sources (Fig. 1a). Other strains belonged to four different minor patterns (X04, X05, X06, and X07); each of these patterns exhibited only two or three genetically related strains.
The present study showed that the PFGE profiles of most of the S. Anatum and S. Enteritidis isolates from food and human stool sources belonged to two clones: X01 and X02. These clones were stable and persisted over a considerable period of time in Tunis. This supports the notion that infected animals and humans are important sources of contamination of the environment and the food chain.
Our findings suggest that certain clones of S. Enteritidis, S. Anatum, S. Corvallis and S. Typhimurium are in circulation in Tunis, suggesting an endemic status for these organisms in Tunisia. PFGE can be regarded as the method most suitable for epidemiological studies. Greater numbers of isolates are needed to evaluate their clonal origins, and the epidemiology of Salmonella in Tunis.
ACKNOWLEDGEMENTS
We thank Dr Fessi Safwan (Service régional de l'hygiène du Ben Arous), the late Dr Chadlia Koubaa (PMI Mellassine), Dr Noureddine Ben Jemmaa (CSB du gouvernorat de Ben Arous), Professor Amel Kechrid (Hôpital d'Enfants, and Professor Taoufik ben Chaâbane (Hôpital la Rabta – Service infectieux) and their staff involved in this study for their assistance with sampling. We thank all public health and hygiene laboratories for their continued cooperation and reporting. We are grateful to Professor Ben Hassen Assia (Centre National de Greffe de Moelle Osseuse, Tunis, Tunisia) for allowing us to use the CHEF DR III apparatus and for providing the E. coli V517 strain. We are grateful to Dr Ben Slama Karim (Laboratoire de Bactériologie, Faculté des Sciences de Tunis, Université Tunis, Tunisia) for providing us with the MVPS 3.31 software.
DECLARATION OF INTEREST
None.







