Human adenoviruses (HAdV), which belong to the genus Mastadenovirus, are grouped into seven species (A–G) that comprise the 55 serologically distinguishable types (designated by Arabic numbers) that have been identified to date [Reference Benkö and Fauquet1–Reference Madisch3]. These viruses have a worldwide distribution and cause a broad spectrum of diseases which can become severe and even fatal in paediatric patients and those with compromised immune systems [Reference Echavarria, Zuckerman, Banatvala and Griffiths4, Reference World, Horwitz, Knipe and Howley5]. A correlation between described subgroups or species and tissue tropism has been described; in general species D (HAdV-8, -19, -37) has been associated with severe keratoconjunctivitis, species B (cluster 2: HAdV-11, -34, -35) with kidney and urinary tract infections; and species A (HAdV-31), F (HAdV-40, -41) and G (HAdV-52) with severe gastroenteritis and diarrhoea [Reference Jones6].
Acute respiratory infection (ARI) is responsible for about 20% of the annual deaths of children aged <5 years worldwide [7] and is frequently associated to adenoviral aetiological agents from species B (cluster 1: HAdV-3, HAdV-7, HAdV-21), C (HAdV-2, HAdV-5) and E (HAdV-4) [Reference Kim8]. In Colombia HAdV is the second most important aetiological viral agent of ARI, after respiratory syncytial virus (RSV), being responsible for about 5% of cases, most of which require hospitalization [Reference Herrera, De la Hoz and Velandia9]. Nonetheless, little is known about the circulating types in the country, in part due to lack of implementation of useful and reliable typing methods. Conventional methods for HAdV typing include culture in cell lines such as A549, HEK293 or HEp-2, specific-sera neutralization or hemagglutination-inhibition assays, processes that are time-consuming and prone to significant reciprocal cross-reactions [Reference Sarantis10]. On the other hand, several molecular approaches have been proposed to circumvent practical problems associated with traditional methods, in which VA or hexon genes are amplified by PCR, followed by type identification by means of size-distinguishable products [Reference Sarantis10], restriction enzyme analysis [Reference Kidd11], or sequencing [Reference Lu and Erman12]. These two genes are excellent molecular targets because they are both highly conserved in all HAdV species; the hexon gene codes for a major virus capsid protein in which type-specific epitopes (targets of neutralizing antibodies in vivo) are encoded by hypervariable regions that allow type differentiation [Reference Lu and Erman12]. The VA gene encodes ‘virus-associated’ (VA) RNAs, which are short non-coding RNA polymerase III transcripts that form secondary structures whose main function is to maintain viral protein expression in infected cells [Reference Coventry and Conn13].
The current study describes the application of a VA gene PCR sequencing approach as an alternative to identify the HAdV circulating types in a Colombian population during 2008. Samples (nasopharyngeal aspirates or swabs) from patients aged <5 years with ARI symptoms (following guidelines established by World Health Organization [7]), were collected in the 18 hospitals and health centres participating in the Respiratory Viruses Sentinel Surveillance Network in Bogota, Colombia. Once collected, the samples were analysed by the Virology Laboratory of the District Health Office (Secretaria Distrital de Salud de Bogota) by immunofluorescence assay (IFA) for the detection of influenza A and B viruses, RSV, parainfluenza 1, 2, and 3 viruses and adenovirus (Light Diagnostics™, Respiratory Panel I Viral Screening and Identification IFA; Millipore, USA). From the 65 samples that tested positive for adenovirus during 2008, 52 were randomly chosen and sent to the Laboratorio de Diagnóstico Molecular y Bioinformática, Universidad de los Andes, where all subsequent molecular analyses were performed. DNA was extracted by using the QIAmp® UltraSens kit (Qiagen Inc., USA), and VA gene amplification was performed by a touchdown PCR using the previously described primers F-VA3a (CGGTSAGGCGYGCGCAGTC), R-VA3b (CGGTAAGACGGGCGCAATC), and R2-VA6 (CGCAGCACNGGATGCATCT) [Reference Kidd11] using the following cycling parameters: an initial denaturing step of 10 min at 95°C, followed by 10 cycles consisting of denaturation at 94°C for 1 min, annealing at 68°C for 1 min (temperature decreased by 1°C every two cycles), and elongation at 72°C for 1 min; 30 cycles of 94°C for 1 min, annealing at 56·5°C for 1 min, and elongation at 72°C for 1 min; with a final elongation step at 72°C for 8 min. A reference influenza B strain, obtained from the National Health Institute (Bogota, Colombia), was included as negative control. PCR products were subjected to electrophoresis on 2% agarose or 8% polyacrylamide, ethidium bromide-containing gels and product purification was performed using the Wizard SV Gel and PCR Clean-Up System (Promega, USA). Positive strand was sequenced by automated sequencing with the PCR forward primer as sequencing primer on an ABI Prism® 310 Genetic Analyzer automatic sequencer (Applied Biosystems, USA). Sequence editing and analysis were performed using Bioedit v. 7.0.9 software [Reference Hall14] and the BLAST Tool from NCBI [Reference Altschul15]. Default parameters for gap opening and extension were used, and the selection of GenBank matching sequences was performed according to the following parameters: E value⩽e−100, highest score value and identity >80%.
The 52 samples included in the study corresponded to patients aged from 1 month to 3 years, most (63·5%) were males aged <1 year (65·4%); these findings were similar to those reported by other studies in Argentina, Cuba and Brazil [Reference Kusznierz16–Reference Moura18]. The most frequent ARI manifestations among all patients corresponded to pneumonia and bronchiolitis, with cough (39/52), fever (36/52), dyspnoea (34/52) and rhinorrhoea (33/52), the most common symptoms.
From the 52 HAdV-positive samples by IFA, 50 were positive by PCR, establishing a 96·15% agreement between the two methods. The two IFA-positive but PCR-negative samples were tested for the presence of PCR inhibitors by amplifying a fragment of human β-globin as described by Saiki et al. [Reference Saiki19]. In both cases the expected 110-bp amplification product was observed confirming the absence of PCR inhibitors, suggesting that the samples could be considered as true PCR-negative, and therefore IFA false-positive, probably due to the subjective nature of immunofluorescence tests. Sample problems such as excessive mucous or technical errors, for example during the washing process could be responsible for this result [Reference Piñón and Savón20].
In 48/50 PCR-positive samples a unique amplicon (500–600 bp) was obtained, corresponding to species with two VA gene copies (HAdV-B cluster 1, HAdV-C, HAdV-D or HAdV-E) whose amplicon size ranged from 400 to 600 bp; in contrast with species with one copy (HAdV-A, HAdV-B cluster 2, or HAdV-F) with amplicon size 240–290 bp [Reference Coventry and Conn13]. Since amplicon size difference is not enough to clearly distinguish the different types, sequencing followed by BLAST analysis was chosen as the typing method. However, in two of the PCR-positive samples the occurrence of two fragments of very similar size (difference <50 bp), most likely due to the presence of more than one type of HAdV in the same sample, made sequencing impossible. For the remaining 48 samples successful identification to the species level was achieved, finding matches with HAdV-B (85·42%, 41/48), C (12·5%, 6/48) and D (2·08%, 1/48). It is of note that this last species had never been reported in Colombia previously, and corresponded to one of the 12 patients who in addition to respiratory symptoms also had conjunctivitis. At the type level, there was only one sample for which the HAdV type could not be elucidated, due to the exact same values for the parameters corresponding to two different HAdV types (HAdV-C6 and -C2) being obtained. This could be explained by the fact that even though there are differences between VA sequences from HAdV types that in most cases allow for differentiation, there are certain types so closely related that their VA sequences are almost identical, which was the case for the two types reported here, as found also by Ma & Mathews [Reference Ma and Mathews21]. In such situations, a second target gene, e.g. hexon, should also be used as described previously [Reference Vabret22]. For the rest of the samples identification was achieved in all cases finding matches with HAdV-1, -2, -3, -5, -6, -7 and -8; with HAdV-3 the most frequent type, found in 22/48 (45·83%) samples (Fig. 1).
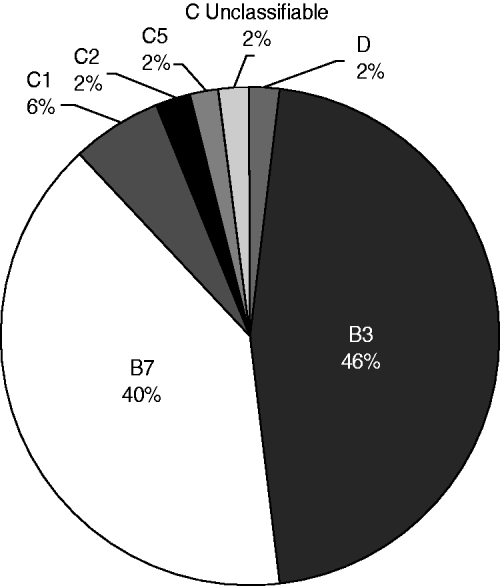
Fig. 1. HAdV types circulating in Bogota from January to November 2008. Unclassifiable refers to the sample in which HAdV type could not be elucidated because the exact same values for the parameters evaluated, corresponding to two different HAdV types (species C, types 6 and 2), were obtained.
As reported in other Latin American countries such as Argentina, Chile and Uruguay, HAdV types 7 and 3 were the predominant circulating types [Reference Kusznierz16, Reference Kajon23]. As described previously by most studies these HAdV types are widely associated with severe ARI manifestations, including bronchiolitis and pneumonia [Reference Kim8, Reference Xu and Erdman24], which were found to be the most frequent manifestations in the patients included in our study. On the other hand, this differs from what has been reported for Brazil, Peru and Mexico, countries in which the most prevalent species is HAdV-C [Reference Moura18, Reference García25, Reference Rosete, Manjarrez and Barrón26]. One of the samples corresponded to HAdV-D (type 8) which is to our knowledge the first report in Colombia of this type in a respiratory sample. Although in general HAdV-D is most commonly associated with eye infections such as severe keratoconjunctivitis, it has already been found in low frequency in respiratory samples in other South American countries, e.g. Argentina [Reference Kusznierz16] and Brazil [Reference Moura18].
Despite the relevance of HAdV as the second most important aetiological agent of ARI in children aged <5 years in Colombia, there is a lack of epidemiological information about these viruses in the country. Reports published to date have focused solely on the detection of the virus with the exception of two studies reporting HAdV typing in Colombia; however, none of them were actually performed in Colombia. In 2007 Herrera-Rodríguez [Reference Herrera-Rodríguez27] published an official report from the National Health Institute (INS, Colombia) that presented results of HAdV typing in samples sent to the Center for Disease Control and Prevention (CDC) in Atlanta. The other study corresponds to a Peruvian study in which samples from several countries were included, of which 27 were Colombian, apparently collected in Medellin [Reference García25]. Therefore, this study is, to the best of our knowledge, the first report of adenovirus circulating types conducted in the country using a molecular approach; and represents an initial effort towards the elucidation of the molecular epidemiology of adenovirus in Colombia. The results presented here show that the protocol described is a useful, rapid and effective approach to HAdV detection and typing in respiratory samples. Nonetheless, due to the limitations of the study mainly concerning sample number, more studies are needed. A larger sample number, from different cities, and for longer periods of time, will allow identification of circulation by season, possible associations with climatic conditions, predominating types in different regions; and possible relationships between types and severity of infection, providing a complete epidemiological profile of respiratory adenoviruses in Colombia.
ACKNOWLEDGEMENTS
We thank medical and paramedical colleagues from Health Centres involved in the Respiratory Viruses Sentinel Surveillance Network in Colombia and the funding provided by the Science Faculty, Universidad de los Andes, Bogota, Colombia.
DECLARATION OF INTEREST
None.



