INTRODUCTION
Recently, a dengue fever (DF) outbreak was suspected in Shenzhen city, China, which appeared to be the first of its kind in the city's history. DF is caused by infection of dengue virus (DENV) – a single-stranded RNA virus of the Flaviviridae family. Based on E-protein antigenic properties, DENV is classified into four serotypes – DENV-1, -2, -3, and -4. DF is a mosquito-transmitted disease, with Aedes aegypti and Aedes albopictus as the main vectors. Despite observation DF epidemics have been more active globally, and no effective DENV vaccine is currently available [Reference Guzmán and Kourí1]. DF epidemics have been particularly prevalent in Southeast Asia, where imported DF cases have caused outbreaks in Guangdong, Hainan, Guangxi, Fujian, Taiwan, Zhejiang, and other southern and southeastern coastal provinces of China. Since the first DF epidemic outbreak occurred in 1978 in Foshan, Guangdong Province, it has increasingly posed a serious public-health threat to China [Reference Wu2].
Shenzhen, a major financial and industrial hub, is one of the largest cities in China. The city has high population mobility due to ever-increasing domestic and international exchanges. Occasionally DF cases have been reported in individuals returning from Southeast Asia and African regions. The climate at this location (22° 32′ 0″ N, 114° 8′ 0″ E) supports A. albopictus reproduction. Although both social and natural factors favour a DF epidemic, a full-blown local outbreak had not occurred previously in Shenzhen.
In October 2010 a group of patients presented with high fever of unknown origin. They were all from an isolated construction site at the mountainous edge of Shenzhen city, and had no recent travel or blood transfusion history. Their clinical manifestations suggested a local DF outbreak. To investigate this epidemic event with serology, molecular biology, and entomology techniques, we obtained blood samples from the patients and their asymptomatic co-workers, and also collected mosquito and larvae samples at the epidemic site. Here we report our results and analysis.
MATERIALS AND METHODS
Specimen source
Starting on 12 October 2010, at a construction site in Futian District, Shenzhen city, a group of nine patients reported sudden symptoms of high fever of unknown origin, head pain, lumbar pain, weakness, chills, skin rash, nausea, vomiting, and other symptoms, which suggested a DF outbreak. Blood samples were collected from the patients, and preserved at −80°C after removing cells by a low-speed centrifugation at 4°C (patient information is detailed in Table 1). Sera of 291 asymptomatic co-workers from the same construction site were also collected, and preserved at −20°C.
Table 1. Summary of suspected patient information and their test results

F, Female; M, male.
Meanwhile, A. albopictus mosquitoes were collected around the construction site. In addition, 9000 A. albopictus larvae were collected at the site, both indoors and outdoors. The larvae were reared to adulthood under standard laboratory conditions (28 ± 2°C at 75–85% relative humidity). The emerged adults were maintained for 3–4 days, sorted for species and gender, pooled (100 individuals/pool), and then stored at –80°C.
Standard DF viral strains and DENV-sensitive cell culture
Standard DENV viral strains (DENV-1, Hawaii strain; DENV-2, New Guinea C strain; DENV-3, H87 strain; DENV-4, H241 strain), C6/36 and BHK-21 cell lines were gifts from the Microbiological Institute of Disease Control and Prevention Center of Guangdong Province. These were stored in liquid nitrogen. After resuscitation, cells were cultured at 28°C or 37°C, plus 5% CO2 in 50% Eagle's minimal essential medium (MEM) +50% RPMI-1640 with 10% fetal bovine serum (containing 100 U/ml penicillin and streptomycin) [Reference Yamada3]. Cells were infected with viruses using conventional methods (see details below) [Reference Yamada3].
Reagents and instruments
MEM, RPMI-1640 and fetal bovine serum were purchased from Gibco (USA); One Step RNA PCR kit (AMV, DRR024A), One Step PrimerScript™ RT–PCR kit (Perfect Real Time, DRR064A), M-MLV enzyme, PrimeSTAR high-fidelity Taq enzyme, and DNA ladder marker (D505A) from Takara Biotechnology Co. (China); the nucleic acid fluorescent PCR kit for DENV (types I–IV) from Shanghai Zhijiang Biological Technology Co. (China); High Pure Viral RNA kit was purchased from Roche (Switzerland); Dengue IgM/IgG Capture ELISA and Dengue Duo Cassette kit from Panbio (Australia); both fluorescent (model no. 7500) and conventional PCR (model no. 9700) machines from ABI (USA); electrophoresis apparatus (basic model) from Bio-Rad (USA).
Serological test
ELISA and immunochromatography assay kits were used to detect DF IgM and IgG antibodies in serum samples according to the manufacturer's instructions (Panbio).
Virus detection and isolation
DENV-sensitive C6/36 and BHK-21 cell lines were used to capture potential viruses in samples [Reference Yamada3]. After growing cells to a monolayer, samples were diluted with maintenance media, and inoculated onto the cells for further culturing. Seven days after inoculation, cytopathic effect (CPE) was examined daily with light microscopy. Culture media were then changed every 7 days, up to 3–5 times. If no CPE was observed, the sample was deemed negative for live viruses. If positive CPE was observed, we harvested the culture media to incubate with fresh cells. This was repeated for several additional passages to confirm a consistent CPE, and also to amplify the virus along the way (continuous passage culturing). At the final passage, if 3/4 cells were CPE positive, the sample was considered positive for live viruses, and the viruses were harvested for further study.
For detecting virus, mosquito samples (males, females, larvae) were homogenized in 1 ml MEM, clarified through a 0·22-mm membrane (Sartorius Biotech GmbH, Germany), and then inoculated onto C6/36 cell cultures for further culturing at 28 °C as described above. All CPE-positive samples were further examined for viral type by RT–PCR (details see below).
DENV nucleic acid detection and type identification
Two hundred μl of serum samples or cell suspensions were processed for RNA extraction with a kit (High Pure Viral RNA kit from Roche). An MGB fluorescent PCR method was used for DENV RNA detection as previously reported, by One Step PrimeScript® RT-PCR kit (Takara Biotechnology) [Reference Liu4]. Dengue RNA was amplified with specific primers: Den-FP (5′-GCATATTGACGCTGGGAGAGA-3′), Den-RP (5′-GGCGTTCTGTGCCTGGAAT-3′), and Den-Probe (5′-CAGAGATCCTGCTGTCTC-MGB 3′) in an ABI7500 machine. When the fluorescence signals were linear, they formed an S-shaped peak against the corresponding cycling data (Ct values ⩽35), which indicated a positive result.
To determine DENV serotype, dengue RNA was amplified using the nested RT–PCR assay described previously [Reference Lanciotti5]. Then, DF virus (types I–IV) nucleic acid was detected (with different primers described above) by the Dengue Virus I–IV Real Time RT–PCR kit according to the manufacturer's instructions (Shanghai ZJ Bio-Tech Co. Ltd, China).
E-gene amplification by RT–PCR
Based on the sequence information of DENV-1 Hawaii strain, we designed two pairs of primers using Primer5 software to cover the full length of the E-gene (Table 1). We purchased the primers from Takara Biotechnology and conducted RT–PCR on the isolated viral samples. The reaction solution consisted of: 28·5 μl ddH2O, 10 μl 5 × buffer, 4 μl dNTP (2·5 mm), 1 μl of each upstream and downstream primer (10 μ m), 5 μl cDNA, and 0·5 μl Taq enzyme. PCR reactions were initiated with a 2-min incubation at 98°C, followed by 30 cycles of 10 s at 98°C, 15 s at 56°C, 60 s at 72°C, with a final 7-min extension at 72°C. The PCR products were separated in 1% agarose gel, analysed with a gel imaging system, and then purified for sequencing.
DNA sequencing and analysis
The purified PCR products of isolated E-gene fragments were sequenced using an ABI373 DNA automatic sequencer (Takara Biotechnology). The nuclear acid positions within each fragment were read at least three times. Sequencing data were assembled and edited electronically using DNAStar software to obtain the full E-gene sequence for each newly isolated virus. Other published DENV sequences were obtained from the GenBank of NCBI (Table 2). The E-gene homology between the isolated virus and other DENV sequences was then analysed using a BLAST program (www.ncbi.nlm.nih.gov/BLAST/). The multiple sequence alignment program Clustal W (version 1.83) was used to obtain an optimal nucleotide sequence alignment file. Next, a phylogenetic tree was reconstructed from the aligned nucleotide sequences following a Neighbour Joining (NJ) P distance method with MEGA v.3.1 software in a Kimura two-parameter model [Reference Goncalvez6]. Bootstrap analysis of 1000 replicates was used to estimate the reliability of the predicted tree.
Table 2. Dengue viral strains used for homology and phylogenetic analysis

Ethical statement
The institutional review board at Shenzhen Centre for Disease Control and Prevention approved the study protocol.
RESULTS
DENV antibodies detected in patients and asymptomatic co-workers
Seven (77·8%) of the nine patients were serum-positive for anti-dengue IgM antibody. Four (44%) of the nine patients were positive for IgG antibody. Fourteen (4·8%) of the 291 asymptomatic co-workers at the construction site were also serum-positive for IgG antibody.
DENV RNA detection and characterization in patients' sera
Eight (88·9%) of the nine patients were serum-positive for DENV RNA via MGB fluorescence PCR assay. We then used RT semi-nested PCR universal primer and type-specific primers to characterize the DENV type. A 511-bp band was detected in patients' sera matching the size of the expected band presented in the DENV-1-positive control sample (Fig. 1). While no band was detected in the negative control sample, smaller bands presented in the positive controls of other DENV types, which did not match the bands detected in patient samples (Fig. 1). In addition, we used the DF virus (types I–IV) nucleic acid fluorescent PCR detection kit to further genotype the isolated strain. We found that the specific primers for DENV-1 generated an obvious peak with an S-shaped curve, while the primers for DENV-2, -3, -4, or the negative control, produced no peak (not shown). These data demonstrate that DENV-1 RNA was present in patients' sera.

Fig. 1. Determination of dengue viral type in serum samples from suspected patients by RT semi-nested PCR. The RT–PCR was performed as described in the text and PCR products were separated in 1% agarose gel and visualized in a gel imaging system. Lanes 1–4, positive controls of representative dengue viral strains: type 1 (Hawaii), type 2 (NGC), type 3 (H87) and type 4 (H241); M, 100-bp DNA ladder marker; lanes 5, 6, 8, patient serum samples; lane 7, negative control.
Virus isolation in patient sera
To examine whether viruses existed in patients' blood, we collected additional serum samples from the eight DENV RNA-positive patients and inoculated viral-sensitive C6/36 or BHK-21 cells. In some inoculated C6/36 cells, we observed characteristic CPE features. These cells began to swell and stick together, the refractive index increased, and intracellular particles accumulated. In BHK-21 cells, CPE appeared at least 5 days later than in C6/36 cells, with signs of focal lesions and aggregations of round, shrunken cells. Continuous passage culturing (see Methods section) further confirmed that three patients positively carried live viruses. All three virus-positive patients were identified using C6/36 cells, but only one was identified using BHK-21 cells. A summary of sample test results of patients is provided in Table 1.
Designation of the currently isolated viral strain as SZ1029
Because all patients were from the same construction site with similar symptoms and the viruses isolated from this epidemic were of the same characteristics (Fig. 1), we initially chose the isolated viruses from one of the patients for further sequencing, and designated it as the SZ1029 strain. That particular patient (donor of no. 1 serum in Table 2) had typical symptoms and was at the earlier stage of the disease. (The SZ1029 strain was isolated from the C6/36 cells inoculated with no. 1 serum.)
E-gene sequencing and homology analysis confirmed SZ1029 was DENV-1
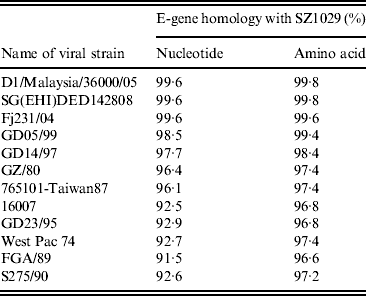
We extracted the SZ1029 viral RNA and amplified it with two pairs of primers corresponding to the E-gene sequence of DENV-1 (Table 3). The end products of 911-bp and 1011-bp fragments matched the expected sizes (data not shown). A complete E-gene sequence was obtained using DNAStar software to combine the sequencing results of the SZ1029 E-gene fragments. The total length was 1485 bp coding 495 amino acids (see Supplementary Tables S1, S2). There was no base deletion or insertion. The SZ1029 nucleotide acid sequence was input into the GenBank database for Blastn search. The SZ1029 sequence has very high similarity to that of known DENV-1 viruses, which further confirms that this epidemic DF was caused by DENV-1. Genetically, SZ1029 is close to the strains reported in Southeast Asia. Of these, SZ1029 has the highest similarity (99·6% homology in nucleotides, 99·8% in amino acids) to the D1/Malaysia/36000/05 (isolated from Malaysia in 2005) [Reference Teoh7] and SG(EHI) DED142808 (isolated from Singapore in 2005) [Reference Lee8] strains. The E-gene nucleotide homology between SZ1029 and the international standard DENV-1 (Hawaii strain) was 94·8%, while their amino acid homology was 97·0%. Comparative analysis with other reported DENV strains showed that the nucleotide homology ranged from 91·5% to 99·6%, while the deduced amino acid sequence homology was from 95·4% to 99·8% (Table 4). The amino acid sequence in E-protein of SZ1029 had two glycosylation loci, which were located at 67–69 (NTT) and 153–155 (NES) sequences of E-protein, respectively. The related virulence loci of DENV-1's E-protein were E44 (E), E156 (T), and E366 (N). There was no variation in the related virulence loci of the SZ1029 strain's E-protein, which is consistent with most DENV-1 strains [Reference Zhao and Yang9]. Furthermore, we sequenced viruses isolated from patient samples nos. 5 and 8. Their E-gene nucleotide sequence data were identical to that of SZ1029, further confirming that the current outbreak was caused by a single viral strain.
Table 3. Primers used for amplifying the dengue E-gene sequence
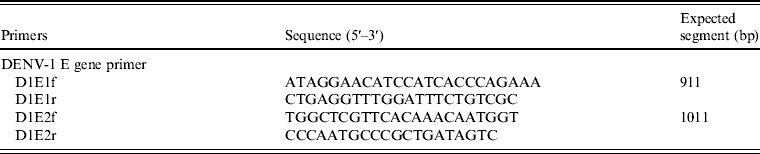
Table 4. Homology comparison of E-gene nucleotide and deduced amino acid sequences between SZ1029 and other known DENV strains
Phylogenetic tree analysis of the SZ1029 strain
In order to further clarify the genetic relationship between the isolated SZ1029 strain and strains reported in other epidemic zones around the world (Table 2), we conducted a phylogenetic analysis. We found that SZ1029 had a very close phylogenetic relationship with the D1/Malaysia/36000/05 and SG(EHI) DED142808 strains, in addition to being close to the Fj231/04, 765101 (Taiwan87), and GZ/80 strains (Fig. 2). According to Gonzalez's classification on gene subtype, both 765101 (Taiwan87) and GZ/80 belong to subtype I. Thus, we assigned SZ1029 to DENV-1.

Fig. 2. Location of the SZ1029 strain in the DENV phylogenetic tree based on E-gene nucleotide sequence. The completed sequence of SZ1029's E-gene was obtained as described in the text. The E-gene sequences of other published DENVs were imported from GeneBank (the information is listed in Table 2). Then a phylogenetic tree was constructed following a Neighbour-Joining method with MEGA v. 3·1 software. The tree topologies were evaluated using 1000 replications of the dataset. The scale bar represents 0·01 nt substitutions/site.
DENV-1 detection in A. albopictus larval samples from the epidemic site
We performed entomological investigations to identify the primary vector at the epidemic site. Initially, we did not detect viruses in the adult mosquito samples collected within and around the construction site, but subsequently detected viruses in mosquito larval samples from the site. Three out of 90 pools displayed CPEs after inoculating C6/36 cells. The RT–PCR experiments further demonstrated that these viruses were DENV-1.
DISCUSSION
We uncovered the first local DF outbreak in Shenzhen city, China from studying the patients, the local population, and the epidemic site. All patients came from the same construction site. They exhibited sudden-onset fever as well as characteristic DF-like pains and rashes for a short period of time (Table 1). None reported recent travel or blood transfusion history. They were serologically positive for DENV-1 antibodies and RNA (Fig. 1, Table 1). Live viruses were isolated from the patients' blood samples and confirmed as DENV-1 by DNA sequencing (Fig. 2, Supplementary Table S1). In addition, viruses were detected in A. albopictus larva samples from the surrounding areas and identified as DENV-1 by PCR. Finally, 4·8% of the asymptomatic co-workers were serologically positive for DENV antibody. These data firmly established that DENV-1 caused this DF outbreak in Shenzhen.
This solitary construction site – now the focal point of the outbreak – lies in the Futian District of Shenzhen, between the edges of Meilin Reservoir and Jiaoye Park in the Meilin Mountain region. The surrounding environment is relatively wild and waterlogged, with many trees and weeds. This is an ideal breeding ground for the DF vector – A. albopictus. The construction site lacked basic mosquito control facilities and procedures and the workers lived in small wooden huts for 1 month prior to the outbreak. All patients reported having been bitten by mosquitoes prior to falling sick. This epidemic emphasizes the importance of preventive measures for workers in mosquito-supporting environments. Because no effective DF vaccine is currently available, mosquito control and prevention measures are our primary means to combat DF epidemics. DF has several phases: incubation, viraemia, and immune response. The available diagnostic tools are differentially sensitive to these stages. To ensure early diagnosis and rapid response, we applied multiple tools in the present study (Table 1).
Up to this time all DF outbreaks in China have been traced back to Southeast Asia [Reference Wu2]. However, in the current study none of the cases clearly identified Southeast Asia as the source of this outbreak. Molecular studies on SZ1029 isolated from this outbreak may provide clues regarding the epidemic's origin. We have focused on the SZ1029 E-protein to provide evidence of origin. E-protein, with 495 amino acids, forms protrusions on the surface of viral particles. This protein determines viral tissue tropism by binding specific host-cell receptors [Reference Moreno-Altamirano, Sánchez-García and Muñoz10], impacts viral virulence, and mediates immunoreactions [Reference Gualano11, Reference Leitmeyer12]. In 2002, Goncalvez et al. established a phylogenetic tree using an adjacent-loci-linking method to analyze the E-protein encoding gene from 44 reported DENV strains [Reference Goncalvez6]. They classified DENV into five genotypes: type I, represented by Hawaii strain (the DENV isolated in 1945); type II includes strains reported during the 1950s–1960s (e.g. the Thailand strain); type III has a single sylvatic strain; type IV includes strains isolated from West Pacific and Australia; and type V are strains mainly isolated in the American continent. Following this example, we constructed a phylogenetic tree to study the genetic relationship between the SZ1029 and other known DENV strains based on their E-gene sequence information (Fig. 2). Our results indicate that SZ1029 has a very close genetic relationship to the D1/Malaysia/36000/05 strain isolated from Malaysia (in 2005), the SG(EHI)DED142808 strain from Singapore (in 2005), and the Fj231/04-strain from Fujian province of China (in 2004), all localizing on the same branch of the phylogenetic tree (Fig. 2). The SZ1029 strain also shares the DENV-1 branch with Hawaii strain, Taiwan87 strain, and GZ/80 strain. Fujian strain-Fj231/04 was confirmed as DENV-1, which was imported by patients initially infected in Southeast Asia [Reference Weng13]. E-protein in SZ1029 has two glycosylation loci, where mutations can affect DENV virulence [Reference Guirakhoo14]. However, E-protein virulence loci of SZ1029 exhibit no variation, consistent with most DENV-1 strains reported in the literature. These loci are relatively conserved in the evolution process of DENV-1. In addition, we studied the E-gene amino acid and nucleotide homology between SZ1029 and other reported strains (Table 4). Our results showed that the E-gene homology between SZ1029 and the DENV-1 international standard (Hawaii strain) is 94·8% in nucleotide sequence. The highest homology was observed with the recently reported D1/Malaysia/36000/05 strain [Reference Teoh7] or SG(EHI)DED142808 strain [Reference Lee8] (both at 99·6% for nucleotide and 99·8% for amino acid). Thus, the potential source of this Shenzhen DF epidemic may originate from Southeast Asia.
However, no patients, their co-workers, or visiting executives, had recently traveled to Southeast Asia. In addition, none had a recent blood transfusion. So what was the transmission mechanism? DF is a mosquito-borne disease. Vertical transmission via a transovarial route in female Aedes mosquitoes occurs in nature. This female-to-offspring mechanism retains the virus for a long time – mosquito eggs can survive in harsh environments for up to a year [Reference Guirakhoo14]. DENV vertical transmission in A. albopictus has been documented both experimentally and in the field [Reference Angel and Joshi15]. Thus, we propose the recent Shenzhen outbreak may have been caused by vertical transmission imported from Southeast Asia: DENV-1-infected female mosquitoes in Southeast Asia laid eggs containing DENV-1, which were transported to the Meilin Mountain of Shenzhen and hatched into adult female mosquitoes still infected with DENV-1, which in turn bit and transmitted the viruses to the construction workers. Indeed, we did detect DENV-1 viruses in some of the mosquito larval samples collected at the epidemic site. This further supports the vertical transmission theory. At present, it is difficult to pinpoint the exact mechanism by which the eggs were imported. But, considering the extensive exchange of agriculture goods, building material, ships, planes, and other materials between Shenzhen and Southeast Asia (transit stops via Hong Kong may also be possible), this transmission route appears to be the most plausible explanation for the recent outbreak.
CONCLUSION
Our study demonstrated that the first DF epidemic in Shenzhen was caused by SZ1029, a DENV-1 strain. Genetic analysis of SZ1029 showed that it had very high homology to the strains recently isolated from Malaysia and Singapore in the DENV-1 evolution branch [Reference Teoh7, Reference Lee8]. Because no imported cases were identified in the Shenzhen outbreak, vertical transmission from mosquito eggs imported from Southeast Asia may be responsible. This trend should be further studied and monitored carefully in the future.
SUPPLEMENTARY MATERIAL
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268813000897.
ACKNOWLEDGEMENTS
This work was supported by Shenzhen Centre for Disease Control and Prevention. C.H.Y. and L.Z. were supported by the Palmer Foundation. We thank Dr. C. Henderson from Henderson Technical Consulting for grammar corrections.
DECLARATION OF INTEREST
None.








