INTRODUCTION
Typhoid fever remains an important disease in developing countries, with an estimated burden of 21·65 million cases and over 200 000 deaths annually worldwide [Reference Crump, Luby and Mintz1]. The epidemiology of the disease is poorly understood in Africa, and there are now constructive efforts to address this [Reference Clemens2]. Parts of South Africa have been identified in the past as highly endemic for the disease [Reference Kustner3], but in recent years, the numbers of typhoid fever cases notified have decreased [4]. Although there are areas that are still endemic for typhoid fever, where there is a lack of potable water and the local population remains at risk; the disease has become uncommon in more urbanized communities, where water treatment plants should supply safe water to local residents.
In 1993, an outbreak of typhoid fever occurred in Delmas, Mpumalanga, a town 100 km due east of Johannesburg, South Africa [Reference Waner5]. More than 1000 cases were admitted to the local field hospital, set up to treat patients affected by the epidemic. Patients from both middle- and low-income groups were affected [Reference Waner5]. The source of the outbreak was postulated to be waterborne, given the numbers of cases that occurred and the high levels of faecal contamination of the local water supply [Reference Waner5].
In September 2005, another outbreak of typhoid fever occurred in the same area. This study was undertaken to document the epidemiology of the outbreak and the likely source of contamination by Salmonella enterica serovar Typhi (S. Typhi). We also compared the molecular epidemiology of the strains from the recent outbreak with those from the previous epidemic to establish whether the two outbreaks were potentially linked.
Pulsed-field gel electrophoresis (PFGE) analysis was employed as our primary genotypic analysis tool to investigate the genetic relatedness of all strains. To augment our PFGE results we used a second genotypic analysis tool, multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA), to further analyse selected strains and use the opportunity to evaluate both techniques in an African context.
MATERIALS AND METHODS
Epidemiological investigation
During September to December 2005, over 600 cases of typhoid fever were reported from the Delmas and Witbank areas of South Africa. A team of epidemiologists from the National Institute for Communicable Diseases (NICD) assisted local health authorities in their investigation by conducting site visits to the area and administering extensive questionnaires regarding exposure to local food and water sources, time spent in the area (local residents vs. visitors), removal of sewerage and mapping these findings on area maps. Due to the numbers of cases, which overwhelmed local health facilities, cases were defined according to the following clinical case definition: pyrexia ⩾38°C for 48 h and a history of living in or travel to the Delmas area.
A case-control investigation was designed based on culture-confirmed cases to establish whether there was an alternative source of infection, besides the water supply. One culture-confirmed case was matched by residence to two controls with the same water source. Variables included age, area of residence, years resident and working in Delmas, exposure to milk and vegetables, history of eating out in the previous 6 weeks, gatherings attended, travel, water volume consumed, availability of a flush toilet, hand washing, access to a piped water supply and whether this was in-house, and usage of water storage containers, as well as the presence or absence of certain clinical features associated with typhoid fever. No history of previous exposure to typhoid fever, prior to this outbreak, was obtained. The statistical analysis was performed at 95% confidence limits. The statistical software package used to analyse the data was Stata 10 (StataCorp., USA). The summary statistic, such as the means for continuous variables and proportions for categorical variables, were provided. Where appropriate, Wilcoxon–Mann–Whitney test or the two-independent sample t test was performed to compare the two groups. Stata's prtesti command was employed to compare the proportions in the two groups.
Ethics
This work received ethical approval from the Health Research Ethics Committee (Human), University of Witwatersrand (clearance number M02-10-42) for the NICD to conduct surveillance and investigate outbreaks of public health importance.
S. Typhi strains
One hundred and three strains of S. Typhi were obtained from the National Health Laboratory Service (NHLS) laboratories serving the affected area at the time of the outbreak for further analysis at the Enteric Diseases Reference Unit (EDRU) of NICD. All strains were biochemically and serologically confirmed as S. Typhi by internationally accepted methods and antimicrobial susceptibilities to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline, nalidixic acid and ciprofloxacin were tested with Etest strips (bioMérieux, France). Extended-spectrum β-lactamase production was tested using the double disk method (MAST Diagnostics, UK). In addition, 43 strains from the Delmas outbreak of 1993 were also available for analysis. A single strain from a stool specimen from an adult female patient in 2007 from the Delmas area was also included in the analysis. An isolate identified on laboratory audit from the same area in a 3-year-old child in December 2008 was not available for analysis, but two further strains from stool specimens from two patients, aged 11 and 13 years, respectively, from the same geographic area in 2009 were also included in the analysis, to make a final total of 149 strains. No history was available for these last four patients.
PFGE analysis
PFGE analysis was performed using a PulseNet standardized protocol [Reference Ribot6] summarized as follows. Bacterial genomic DNA was digested with XbaI restriction endonuclease (Roche Diagnostics GmbH, Germany). For control purposes, a strain of S. Braenderup (strain H9812) was included as a reference standard and analysed in parallel with all typhoid strains. Digested DNA was separated on a 1% agarose gel (SeaKem Gold agarose, USA) using a CHEF-DR III electrophoresis system (Bio-Rad Laboratories Inc., USA) programmed with an electrophoresis gradient of 6 V/cm, an included angle of 120°, an initial switch time of 2·2 s, a final switch time of 63·8 s and a run time of 19 h. Following electrophoresis, agarose gels were stained with ethidium bromide and patterns were visualized following UV illumination. Images of the patterns were captured into BioNumerics (version 6.01) software (Applied Maths, Belgium) for further analysis and comparison. All test patterns were normalized against the pattern of the S. Braenderup reference standard. Dendrograms of the patterns were created using the unweighted pair-group method with arithmetic averages, with analysis of banding patterns incorporating the Dice coefficient at an optimization setting of 0·5% and a position tolerance setting of 1·5%. A cluster of strains was defined as ⩾4 strains with patterns having ⩾98% similarity on the dendrogram.
Preparation of crude bacterial DNA
From 2 to 4 colonies of bacteria cultured on 5% blood agar (Diagnostic Media Products, South Africa) was resuspended in 400 μl of 10 mm Tris–1 mm EDTA buffer (pH 8) and boiled at 95°C for 20 min. The suspension was then centrifuged at 6000 rpm for 3 min and the resulting supernatant (crude DNA preparation) was used as a template for PCR.
MLVA
MLVA was based on three VNTR gene loci (TR1, TR2, TR3) as previously described [Reference Liu7]. This method employed manual agarose gel electrophoresis analysis, but was modified to incorporate automated capillary electrophoresis of fluorescently labelled PCR products. The make-up of PCR primers used to amplify VNTR loci are shown in Table 1. The forward primer for each locus was labelled with a distinctive fluorescent dye (Applied Biosystems, USA). Each VNTR locus was amplified in a separate PCR of 25 μl final volume containing 1 μl crude bacterial DNA, 2 mm MgCl2, 0·5 μm of each primer, 200 μm deoxynucleotide triphosphates (Bioline, UK), 1 U AmpliTaq Gold DNA polymerase (Applied Biosystems) and 1× AmpliTaq Gold DNA polymerase buffer (Applied Biosystems); with thermal cycling (25 times) at 95°C for 75 s, 55°C for 75 s and 72°C for 75 s. The three resultant PCRs were pooled as follows: 2 μl TR1+2 μl TR2+6 μl TR3. This pooled mixture was then diluted 1:40 in deionized water. Two microlitres of this diluted mixture was then mixed with 0·5 μl GeneScan 600 LIZ size standard (Applied Biosystems) and 7·5 μl Hi-Di formamide (Applied Biosystems). This 10-μl mixture was then incubated at 95°C for 3 min and cooled to room temperature before being subjected to capillary electrophoresis using an Applied Biosystems 3130 Genetic Analyzer. Electrophoresis was performed through POP-7 polymer (Applied Biosystems) at 15 kV for 25 min at a temperature of 60°C. Raw data was captured and analysed using GeneMapper (version 4.0) software (Applied Biosystems) which identified each VNTR locus by its distinctive colour (fluorescence) and automatically sized the gene product via comparison to the internal size standard.
Table 1. PCR primers used for MLVA

RESULTS
Epidemiological investigation
We identified 49 cases and 96 controls for the final analysis. Cases were clustered in certain areas of the town. As the outbreak evolved, it spread from the more affluent areas (Delmas) to the neighbouring areas (Botleng and Botleng extensions), these areas eventually having the greatest burden of disease (Fig. 1). There were no statistical differences in exposure of cases and controls to potential sources of infection, including food outlets for fresh and pre-prepared food, various water sources (tap, wells, ground water, bottled water), toilet facilities and hand-washing practices at work and home.
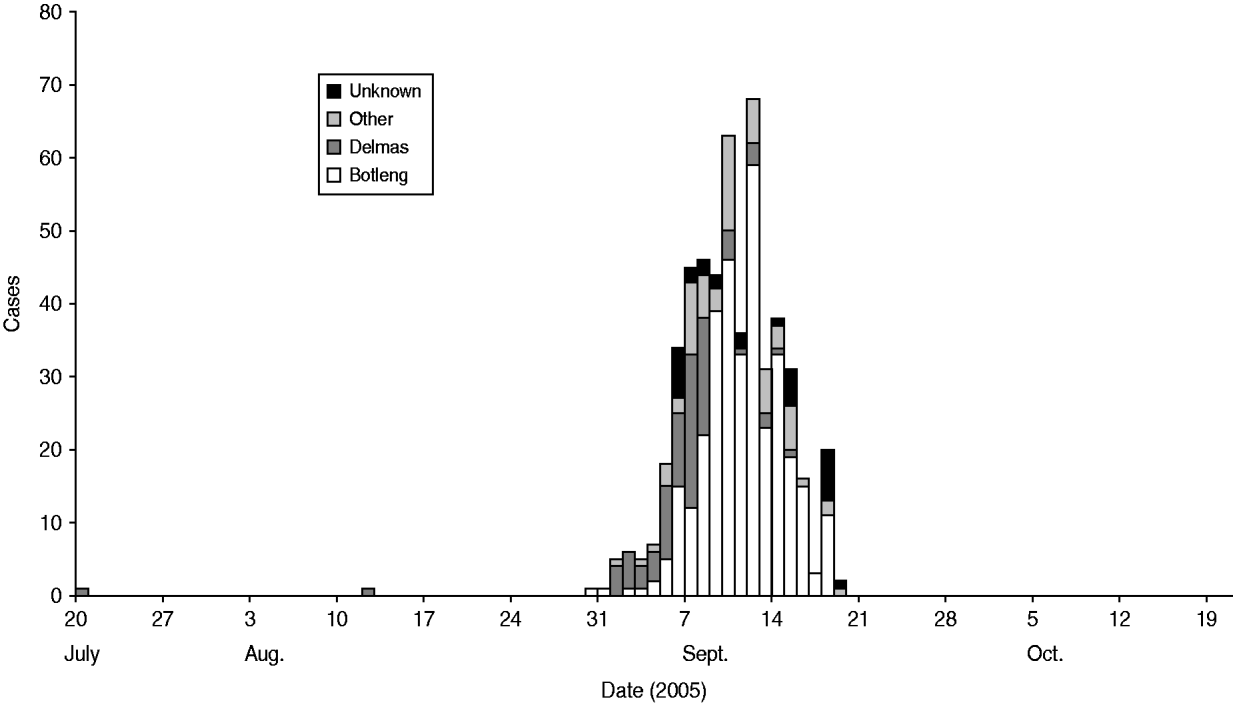
Fig. 1. Suspected typhoid cases by place of residence, Delmas, 2005.
The minimum and maximum ages of the controls and cases were 2 years and 67 years, and 1 year and 62 years, respectively, i.e. the age ranges for controls and cases were 65 years and 61 years, respectively. Controls were significantly older than cases (controls: mean age 32·58 years; cases: mean age 20·97 years; P=0·003).
A significantly greater proportion of controls had contact with someone with symptoms of typhoid fever than cases [81/96 (84%) vs. 33/49 (67%); z=−2·3656; P=0·018]. Therefore, cases were less likely to have had contact with someone with symptoms of typhoid fever. Significantly, a larger proportion of controls attended a gathering than cases [49/96 (51%) vs. 16/49 (33%); z=2·1060, P=0·035]. Clearly, cases were less likely to have attended any form of gathering.
There was no statistically significant difference between cases and controls in either the number of years working (z=1·339, P=0·1807) or living (z=−0·438, P=0·6613) in Delmas, or in number of glasses (quantity) of water drunk per day (z=0·095, P=0·9244).
S. Typhi strains
All isolates submitted for testing to EDRU were confirmed as S. Typhi and were susceptible to all antibiotics tested.
PFGE
PFGE analysis was performed on 149 S. Typhi strains from the Delmas area between 2005 and 2009: this revealed a diversity of 53 unique PFGE patterns. Dendrogram analysis (see Supplementary Fig. 1, available online) of PFGE patterns revealed six clusters (A–F). Strains from the 1993 outbreak were found in clusters A, B and E; while strains from the 2005 outbreak were found in all six clusters. Clusters A, B and E were represented by strains from both the 1993 and 2005 outbreaks. Clusters C, D and F represented infections from 2005. Cluster B was of particular interest as it was our largest cluster representing 30% (44/149) of our strains and in addition to its 1993 and 2005 representation, it also included a single strain from 2007. Cluster C was of particular interest as it included two strains from 2009.
MLVA
MLVA results are reported as MLVA types which represent MLVA allele profiles defining the size of PCR products for three gene loci in the order TR1, TR2 and TR3. Each unique MLVA allele profile was assigned a specific MLVA type number.
Example 1
MLVA type 5 represented the MLVA profile ‘254-403-422’ which defined a 254-bp product for the TR1 locus, a 403-bp product for the TR2 locus and a 422-bp product for the TR3 locus.
Example 2
MLVA profile ‘254 ___ 422’ indicated a profile with no amplification of PCR product at the TR2 locus.
MLVA was performed on 149 S. Typhi strains from the Delmas area between 2005 and 2009, revealing a diversity of 49 unique MLVA types (Supplementary Fig. 1). MLVA types 1, 5 and 7 were the most frequently encountered, with MLVA type 1 accounting for 24% (35/149) of the strains, MLVA type 5 accounting for 13% (20/149) of the strains and MLVA type 27 accounting for 10% (15/149) of the strains. Sixteen MLVA types were unique to strains from 1993, while 28 MLVA types were unique to strains from 2005. Only four MLVA types (1, 36, 37, 48) were found among strains from both the years 1993 and 2005. In addition, MLVA type 1 was also the MLVA type for the single strain from 2007 and also the MLVA type for one of the strains from 2009.
DISCUSSION
An extensive epidemiological investigation, including both a case-control study as well as follow-up of clinically diagnosed cases of this outbreak supported the premise that it was the result of contaminated water supplies to the town. The absence of other common exposures, such as food outlets, common gatherings or occupational exposure, confirmed that these potential sources of typhoid fever could be excluded. The water reticulation system in the town has been previously examined [Reference Waner5], but has remained unchanged since the previous typhoid fever outbreak in the area in 1993. Exposure to different water sources was not significantly different between cases and controls, but clustering of cases in certain parts of the town and the relationship of these clusters to the molecular clusters, could be related to water reticulation patterns, supporting the premise that water was the primary vehicle in this outbreak. Exposure to commercial food outlets in the area was not significantly different between cases and controls, but attending gatherings appeared to be protective, implying that exposure to commercial food outlets was unlikely to be a vehicle in this instance. Exposure to patients with symptoms consistent with typhoid fever also appeared protective, negating a role for person-to-person transmission. The only other significant difference that could be found between cases and controls was in the median age, which would support the suggestion that the older members of the community may have been exposed to typhoid fever in 1993 and had retained their immunity. The typhoid fever outbreak in Delmas at the time was associated with an outbreak of gastroenteritis, and we believe, given the time interval of 12 years, that all patients were treated at the same field site and that these patients were primarily from disadvantaged socioeconomic backgrounds [Reference Waner5], recall bias may have affected responses to questions regarding previous exposures [Reference Kroeger8, Reference Manesh9]. As this information was not sought in the questionnaire, we cannot conclusively support this, but given the likely exposure to contaminated water, it is a reasonable explanation.
Molecular epidemiological (both PFGE analysis and MLVA investigations) methods suggested that the 1993 outbreak could be related to the later one in 2005, as well as to isolated cases of typhoid fever occurring in the Delmas area subsequently. PFGE results confirmed that strains from the 1993 outbreak were found in clusters A, B and E; while strains from the 2005 outbreak were found in all six clusters. As clusters A, B and E were represented by strains from both the 1993 and 2005 outbreaks, these strains would have caused infection in 1993 and then reappeared to cause an outbreak again in 2005. Clusters C, D and F were not represented by any strains from the 1993 outbreak and therefore could represent new infections in 2005 or mutations to the genome of the original strains from the 1993 outbreak. MLVA type 1 was the foremost MLVA type among all strains and may have been the index MLVA type for typhoid fever in Delmas which originated in 1993, reappeared in 2005 and still persisted in 2007 and 2009.
We used the opportunity to evaluate two molecular epidemiological methods for S. Typhi, but we cannot conclude, with these results, that one method is better than the other. For our 149 strains, PFGE analysis showed 53 unique PFGE patterns, while MLVA showed 49 unique MLVA types. Both methods were useful and the results of one complemented the results of the other, although this lengthened and complicated the protocols for optimizing molecular methods in an epidemiological investigation. For example, strains within PFGE cluster B could be separated into 10 MLVA types (1–5, 12, 27, 37, 54, 55) which would suggest that MLVA has the better discriminatory ability. However, strains with MLVA type 1 could be separated into eight PFGE patterns located in five clusters (clusters A, B, C, D, F) which implied that PFGE may have the better discriminatory ability. This shows that both methods provided value to our analysis and should enjoy equal status. The major advantage of MLVA is that results could be obtained within a day compared to the 3 days required for PFGE analysis, although PFGE is highly standardized and can be done in the absence of sequencing information of the bacterial genome. There may thus be potential to optimize MLVA further or develop alternative molecular epidemiological methods to better evaluate potential S. Typhi outbreaks.
Ongoing long-term exposure in an endemic area for typhoid fever has previously been associated with the development of protective levels of antibodies to the Vi capsular antigen, the major component of the currently available parenteral vaccine for typhoid fever, in unvaccinated individuals [Reference Keddy10]. This suggests that S. Typhi, through the presence of an undiagnosed carrier or carriers and possibly other bacteria with common antigens continued to circulate in the area for 12 years, resulting in the protection of older individuals in Delmas. When the system for the provision of safe water failed again in 2005 [Reference Sidley11], this allowed environmental contamination of the ground water/borehole system and the second outbreak occurred in a younger, previously unexposed population.
A recommendation was issued through NICD to treat all patients with clinically suspected typhoid fever with ciprofloxacin, including in the absence of laboratory-confirmed diagnosis, because of ease of administration, the shorter treatment period required and the available information on antimicrobial susceptibility. We believe that in this outbreak it was the optimal treatment, as generally response to therapy was good and there were no recorded treatment failures. A long-term carrier state for S. Typhi or treatment failures appear to be rare in patients treated with ciprofloxacin and this may have contributed in part to controlling the outbreak, in the absence of a permanent solution to the level of water contamination [Reference Sidley11, Reference Nguyen12].
It is a point of concern for us that typhoid fever was diagnosed in a 3-year-old child in 2008, born after the 2005 outbreak, from whom the isolate was unavailable for analysis. Additionally, there were three further isolates from patients in the Delmas area from 2007 and 2009: these isolates of S. Typhi showed PFGE patterns indistinguishable to patterns associated with the 1993 and 2005 outbreaks. With regard to the latter three cases, the single strain from 2007 showed a PFGE pattern correlating with cluster B, while two strains from 2009 showed a PFGE pattern correlating with cluster C. In addition, the single strain from 2007 and one of the strains from 2009, corresponded to MLVA type 1, the foremost MLVA type associated with strains from 1993 and 2005. As no further history or additional epidemiological information was available for these four patients, we cannot say whether these isolates were associated with carriage following clinical disease. However, this suggests, that the strains of S. Typhi that caused the recent outbreak may still be circulating in the area, in association with a carrier or carriers, either in local water supplies or through environmental contamination by an unidentified carrier.
Due to financial constraints, not all cases diagnosed clinically as typhoid fever were followed up for carriage in either 1993 [Reference Waner5] or 2005 (B. N. Harris, personal communication). The Delmas community again was affected by extensive outbreaks of gastroenteritis in 2007 and 2009 [13, 14], again calling into question water quality. It is possible that after two outbreaks, immunity to typhoid fever in the community is high, and thus no further outbreaks have occurred in association with the outbreaks of gastroenteritis. As S. Typhi appears to be still present in the community, associated with isolated cases, further outbreaks may occur if urgent and appropriate steps are not taken to ensure that inhabitants of Delmas have safe water.
NOTE
Supplementary material accompanies this paper on the Journal's website (http://journals.cambridge.org/hyg).
ACKNOWLEDGEMENTS
We thank Mrs L. De Gouveia for supplying us with S. Typhi strains from the first outbreak in Delmas in 1993. This work was supported by a grant by the NHLS Research Trust. S. Typhi isolates were collected through the Group for Enteric, Respiratory and Meningeal Disease Surveillance Network, South Africa.
DECLARATION OF INTEREST
None.




